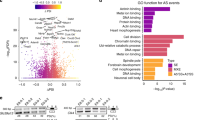
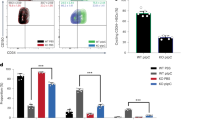
Abstract
Previous investigations of the core gene regulatory circuitry that controls the pluripotency of embryonic stem (ES) cells have largely focused on the roles of transcription, chromatin and non-coding RNA regulators1,2,3. Alternative splicing represents a widely acting mode of gene regulation4,5,6,7,8, yet its role in regulating ES-cell pluripotency and differentiation is poorly understood. Here we identify the muscleblind-like RNA binding proteins, MBNL1 and MBNL2, as conserved and direct negative regulators of a large program of cassette exon alternative splicing events that are differentially regulated between ES cells and other cell types. Knockdown of MBNL proteins in differentiated cells causes switching to an ES-cell-like alternative splicing pattern for approximately half of these events, whereas overexpression of MBNL proteins in ES cells promotes differentiated-cell-like alternative splicing patterns. Among the MBNL-regulated events is an ES-cell-specific alternative splicing switch in the forkhead family transcription factor FOXP1 that controls pluripotency9. Consistent with a central and negative regulatory role for MBNL proteins in pluripotency, their knockdown significantly enhances the expression of key pluripotency genes and the formation of induced pluripotent stem cells during somatic cell reprogramming.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Data deposits
GEO accession numbers are provided in Supplementary Table 1.
References
Young, R. A. Control of the embryonic stem cell state. Cell 144, 940–954 (2011)
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012)
Bao, X. et al. MicroRNAs in somatic cell reprogramming. Curr. Opin. Cell Biol. 25, 208–214 (2013)
Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genet. 40, 1413–1415 (2008)
Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008)
Braunschweig, U., Gueroussov, S., Plocik, A. M., Graveley, B. R. & Blencowe, B. J. Dynamic integration of splicing within gene regulatory pathways. Cell 152, 1252–1269 (2013)
Nilsen, T. W. & Graveley, B. R. Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463 (2010)
Kalsotra, A. & Cooper, T. A. Functional consequences of developmentally regulated alternative splicing. Nature Rev. Genet. 12, 715–729 (2011)
Gabut, M. et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell 147, 132–146 (2011)
Chen, X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008)
Kim, J., Chu, J., Shen, X., Wang, J. & Orkin, S. H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 (2008)
Silva, J. et al. Nanog is the gateway to the pluripotent ground state. Cell 138, 722–737 (2009)
Irimia, M. & Blencowe, B. J. Alternative splicing: decoding an expansive regulatory layer. Curr. Opin. Cell Biol. 24, 323–332 (2012)
Rao, S. et al. Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol. Cell. Biol. 30, 5364–5380 (2010)
Salomonis, N. et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc. Natl Acad. Sci. USA 107, 10514–10519 (2010)
Mayshar, Y. et al. Fibroblast growth factor 4 and its novel splice isoform have opposing effects on the maintenance of human embryonic stem cell self-renewal. Stem Cells 26, 767–774 (2008)
Barash, Y. et al. Deciphering the splicing code. Nature 465, 53–59 (2010)
Liang, J. et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nature Cell Biol. 10, 731–739 (2008)
Lian, I. et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 24, 1106–1118 (2010)
Wang, E. T. et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 150, 710–724 (2012)
Charizanis, K. et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron 75, 437–450 (2012)
Pascual, M., Vicente, M., Monferrer, L. & Artero, R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation 74, 65–80 (2006)
Licatalosi, D. D. et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469 (2008)
Fernandez-Costa, J. M., Llamusi, M. B., Garcia-Lopez, A. & Artero, R. Alternative splicing regulation by Muscleblind proteins: from development to disease. Biol. Rev. Camb. Philos. Soc. 86, 947–958 (2011)
Woltjen, K. et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458, 766–770 (2009)
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007)
Golipour, A. et al. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell 11, 769–782 (2012)
Samavarchi-Tehrani, P. et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77 (2010)
Calarco, J. A. et al. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 21, 2963–2975 (2007)
Labbé, R. M. et al. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells 30, 1734–1745 (2012)
Gabut, M. et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell 147, 132–146 (2011)
Polo, J. M. & Hochedlinger, K. When fibroblasts MET iPSCs. Cell Stem Cell 7, 5–6 (2010)
Samavarchi-Tehrani, P. et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77 (2010)
Golipour, A. et al. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell 11, 769–782 (2012)
Hotta, A. et al. EOS lentiviral vector selection system for human induced pluripotent stem cells. Nature Protocols 4, 1828–1844 (2009)
Hotta, A. et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nature Methods 6, 370–376 (2009)
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007)
Cheung, A. Y. et al. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum. Mol. Genet. 20, 2103–2115 (2011)
Labbé, R. M. et al. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells 30, 1734–1745 (2012)
Xiong, H. Y., Barash, Y. & Frey, B. J. Bayesian prediction of tissue-regulated splicing using RNA sequence and cellular context. Bioinformatics 27, 2554–2562 (2011)
Wang, E. T. et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 150, 710–724 (2012)
Licatalosi, D. D. et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469 (2008)
Irimia, M., Rukov, J. L., Roy, S. W., Vinther, J. & Garcia-Fernandez, J. Quantitative regulation of alternative splicing in evolution and development. Bioessays 31, 40–50 (2009)
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2009)
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009)
Acknowledgements
The authors thank U. Braunschweig, J. Ellis, S. Gueroussov and B. Raj for comments on the manuscript. We acknowledge D. Torti in the Donnelly Sequencing Centre for sequencing samples; L. Lee for assisting with the splicing code analysis; J. Garner (Hospital for Sick Children Embryonic Stem Cell Facility) for preparing feeder cells; A. Piekna for morphological examination of human iPSC colonies; M. Narimatsu for assisting with chimaerism analysis; and P. Mero for assisting with cell imaging. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) (to B.J.B., J.L.W., A.N., J.E. and B.J.F.), the Ontario Research Fund (to J.L.W., B.J.B., A.N. and others), the Canadian Stem Cell Network (to A.N. and B.J.B.), and by a grant from the National Institutes of Health (R33MH087908) to J.E. H.H. was supported by a University of Toronto Open Fellowship. P.J.R., M.I. and N.L.B.-M. were supported by postdoctoral fellowships from the Ontario Stem Cell Initiative, Human Frontiers Science Program Organization, and the Marie Curie Actions, respectively.
Author information
Authors and Affiliations
Contributions
H.H. performed experiments in Figs 1–4 and Supplementary Figs 2–9 and 11–13. M.I. performed bioinformatic analyses in Figs 1–4 and Supplementary Figs 1, 3, 7, 10 and 14, with input from N.L.B.-M. L.D. and A.G. assisted with secondary MEF reprogramming experiments and clone characterization, and D.T. generated secondary MEF lines and performed chimaerism testing. P.J.R., T.T. and M.G. performed human reprogramming experiments and iPSC characterization. H-K.S. performed teratoma assays. B.A. and B.J.F. generated splicing code data. I.P.M., H.-K.S. and D.O. assisted with ES-cell overexpression and differentiation experiments. E.W. and C.B.B. generated and analysed CLIP-seq data. E.N.N. and V.S. performed RT–PCR validation experiments. B.J.B., H.H. and M.I. designed the study, with input from J.L.W., J.E., A.N. and J.M. B.J.B., H.H. and M.I. wrote the manuscript, with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary References, full legends for Supplementary Tables 1-5 and Supplementary Figures 1-15. (PDF 8925 kb)
Supplementary Table 1
This file contains information on RNA-Seq datasets and samples – see Supplementary Information for full legend. (XLSX 48 kb)
Supplementary Table 2
This file contains information on Human and mouse ESC-differential AS events - Supplementary Information for full legend. (XLSX 195 kb)
Supplementary Table 3
This file contains DAVID (http://david.abcc.ncifcrf.gov/) output for functional enrichment categories for human, mouse or conserved ESC-differential AS events – see Supplementary Information for full legend. (XLSX 168 kb)
Supplementary Table 4
This file contains expression levels of the human and mouse splicing factors analyzed by RNA-Seq - see Supplementary Information for full legend. (XLSX 199 kb)
Supplementary Table 5
This file contains information on mouse ESC-differential AS events plotted in Supplementary Figure 14- see Supplementary Information for full legend. (XLSX 63 kb)
Rights and permissions
About this article
Cite this article
Han, H., Irimia, M., Ross, P. et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 498, 241–245 (2013). https://doi.org/10.1038/nature12270
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12270
This article is cited by
-
Molecular versatility during pluripotency progression
Nature Communications (2023)
-
The physiology of alternative splicing
Nature Reviews Molecular Cell Biology (2023)
-
TEQUILA-seq: a versatile and low-cost method for targeted long-read RNA sequencing
Nature Communications (2023)
-
RBFOX2 deregulation promotes pancreatic cancer progression and metastasis through alternative splicing
Nature Communications (2023)
-
RSL24D1 sustains steady-state ribosome biogenesis and pluripotency translational programs in embryonic stem cells
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.