Abstract
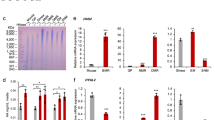
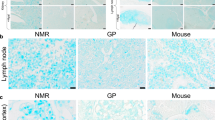
The naked mole rat (Heterocephalus glaber) displays exceptional longevity, with a maximum lifespan exceeding 30 years1,2,3. This is the longest reported lifespan for a rodent species and is especially striking considering the small body mass of the naked mole rat. In comparison, a similarly sized house mouse has a maximum lifespan of 4 years4,5. In addition to their longevity, naked mole rats show an unusual resistance to cancer. Multi-year observations of large naked mole-rat colonies did not detect a single incidence of cancer2,6. Here we identify a mechanism responsible for the naked mole rat’s cancer resistance. We found that naked mole-rat fibroblasts secrete extremely high-molecular-mass hyaluronan (HA), which is over five times larger than human or mouse HA. This high-molecular-mass HA accumulates abundantly in naked mole-rat tissues owing to the decreased activity of HA-degrading enzymes and a unique sequence of hyaluronan synthase 2 (HAS2). Furthermore, the naked mole-rat cells are more sensitive to HA signalling, as they have a higher affinity to HA compared with mouse or human cells. Perturbation of the signalling pathways sufficient for malignant transformation of mouse fibroblasts fails to transform naked mole-rat cells. However, once high-molecular-mass HA is removed by either knocking down HAS2 or overexpressing the HA-degrading enzyme, HYAL2, naked mole-rat cells become susceptible to malignant transformation and readily form tumours in mice. We speculate that naked mole rats have evolved a higher concentration of HA in the skin to provide skin elasticity needed for life in underground tunnels. This trait may have then been co-opted to provide cancer resistance and longevity to this species.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
17 July 2013
A minor change was made to the Fig. 4b legend.
References
Buffenstein, R. & Jarvis, J. U. The naked mole rat—a new record for the oldest living rodent. Sci. Aging Knowl. Environ. 2002, pe7 (2002)
Buffenstein, R. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J. Comp. Physiol. B 178, 439–445 (2008)
Kim, E. B. et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479, 223–227 (2011)
Turturro, A. et al. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J. Gerontol. A 54, B492–B501 (1999)
de Magalhaes, J. P., Costa, J. & Toussaint, O. HAGR: the Human Ageing Genomic Resources. Nucleic Acids Res. 33, D537–D543 (2005)
Delaney, M. A., Nagy, L., Kinsel, M. J. & Treuting, P. M. Spontaneous histologic lesions of the adult naked mole rat (Heterocephalus glaber): A retrospective survey of lesions in a zoo population. Vet. Pathol.. http://dx.doi.org/10.1177/0300985812471543 (2013)
Seluanov, A. et al. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc. Natl Acad. Sci. USA 106, 19352–19357 (2009)
Abercrombie, M. Contact inhibition and malignancy. Nature 281, 259–262 (1979)
Toole, B. P. Hyaluronan: from extracellular glue to pericellular cue. Nature Rev. Cancer 4, 528–539 (2004)
Kothapalli, D. et al. Hyaluronan and CD44 antagonize mitogen-dependent cyclin D1 expression in mesenchymal cells. J. Cell Biol. 176, 535–544 (2007)
Puré, E. & Assoian, R. K. Rheostatic signaling by CD44 and hyaluronan. Cell. Signal. 21, 651–655 (2009)
Holmes, M. W., Bayliss, M. T. & Muir, H. Hyaluronic acid in human articular cartilage. Age-related changes in content and size. Biochem. J. 250, 435–441 (1988)
Jiang, D., Liang, J. & Noble, P. W. Hyaluronan in tissue injury and repair. Annu. Rev. Cell Dev. Biol. 23, 435–461 (2007)
Watanabe, K. & Yamaguchi, Y. Molecular identification of a putative human hyaluronan synthase. J. Biol. Chem. 271, 22945–22948 (1996)
Stern, R. & Jedrzejas, M. J. Hyaluronidases: their genomics, structures, and mechanisms of action. Chem. Rev. 106, 818–839 (2006)
Ponta, H., Sherman, L. & Herrlich, P. A. CD44: from adhesion molecules to signalling regulators. Nature Rev. Mol. Cell Biol. 4, 33–45 (2003)
Morrison, H. et al. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 15, 968–980 (2001)
Hahn, W. C. et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 22, 2111–2123 (2002)
Rangarajan, A., Hong, S. J., Gifford, A. & Weinberg, R. A. Species- and cell type-specific requirements for cellular transformation. Cancer Cell 6, 171–183 (2004)
Liang, S., Mele, J., Wu, Y., Buffenstein, R. & Hornsby, P. J. Resistance to experimental tumorigenesis in cells of a long-lived mammal, the naked mole-rat (Heterocephalus glaber). Aging Cell 9, 626–635 (2010)
Lee, H. G. & Cowman, M. K. An agarose gel electrophoretic method for analysis of hyaluronan molecular weight distribution. Anal. Biochem. 219, 278–287 (1994)
Acknowledgements
This work was supported by the grants from the US National Institutes of Health and Ellison Medical Foundation to V.G. We thank M. Van Meter for critically reading the manuscript.
Author information
Authors and Affiliations
Contributions
X.T. performed HA analysis, HAase assays, soft agar assays, and generated cells for xenograft experiments; J.A. performed immunoblots and cloning and analysis of HAS2; C.H. identified HA, performed tissue staining, and soft agar assays; A.V. performed xenografts; M.-M.R. performed HA affinity assays; J.A. purified HA; Z.M. performed experiments with HAS2 expression; E.N. provided essential materials; X.T., J.A., C.H., A.S. and V.G. designed the study and analysed data; A.S. and V.G. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-9. (PDF 7768 kb)
Rights and permissions
About this article
Cite this article
Tian, X., Azpurua, J., Hine, C. et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499, 346–349 (2013). https://doi.org/10.1038/nature12234
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12234
This article is cited by
-
Aging research comes of age
Nature Methods (2024)
-
Experimental evidence for cancer resistance in a bat species
Nature Communications (2024)
-
Increased hyaluronan by naked mole-rat Has2 improves healthspan in mice
Nature (2023)
-
miRNA-214-3p stimulates carcinogen-induced mammary epithelial cell apoptosis in mammary cancer-resistant species
Communications Biology (2023)
-
Postnatal oogenesis leads to an exceptionally large ovarian reserve in naked mole-rats
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.