Abstract
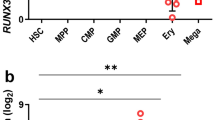
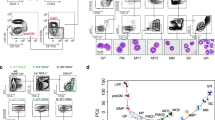
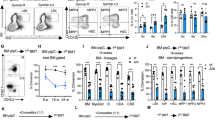
Stem cells and progenitors in many lineages undergo self-renewing divisions, but the extracellular and intracellular proteins that regulate this process are largely unknown. Glucocorticoids stimulate red blood cell formation by promoting self-renewal of early burst-forming unit–erythroid (BFU–E) progenitors1,2,3,4. Here we show that the RNA-binding protein ZFP36L2 is a transcriptional target of the glucocorticoid receptor (GR) in BFU–Es and is required for BFU–E self-renewal. ZFP36L2 is normally downregulated during erythroid differentiation from the BFU–E stage, but its expression is maintained by all tested GR agonists that stimulate BFU–E self-renewal, and the GR binds to several potential enhancer regions of ZFP36L2. Knockdown of ZFP36L2 in cultured BFU–E cells did not affect the rate of cell division but disrupted glucocorticoid-induced BFU–E self-renewal, and knockdown of ZFP36L2 in transplanted erythroid progenitors prevented expansion of erythroid lineage progenitors normally seen following induction of anaemia by phenylhydrazine treatment. ZFP36L2 preferentially binds to messenger RNAs that are induced or maintained at high expression levels during terminal erythroid differentiation and negatively regulates their expression levels. ZFP36L2 therefore functions as part of a molecular switch promoting BFU–E self-renewal and a subsequent increase in the total numbers of colony-forming unit–erythroid (CFU–E) progenitors and erythroid cells that are generated.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Accessions
Gene Expression Omnibus
Data deposits
All microarray data are available from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under accession code GSE46216.
References
Flygare, J., Rayon Estrada, V., Shin, C., Gupta, S. & Lodish, H. F. HIF1α synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood 117, 3435–3444 (2011)
Bauer, A. et al. The glucocorticoid receptor is required for stress erythropoiesis. Genes Dev. 13, 2996–3002 (1999)
Wessely, O., Deiner, E. M., Beug, H. & von Lindern, M. The glucocorticoid receptor is a key regulator of the decision between self-renewal and differentiation in erythroid progenitors. EMBO J. 16, 267–280 (1997)
Reichardt, H. M. et al. DNA binding of the glucocorticoid receptor is not essential for survival. Cell 93, 531–541 (1998)
Richmond, T. D., Chohan, M. & Barber, D. L. Turning cells red: signal transduction mediated by erythropoietin. Trends Cell Biol. 15, 146–155 (2005)
Wu, H., Liu, X., Jaenisch, R. & Lodish, H. F. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell 83, 59–67 (1995)
Hattangadi, S. M., Wong, P., Zhang, L., Flygare, J. & Lodish, H. F. From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood 118, 6258–6268 (2011)
Narla, A. et al. Dexamethasone and lenalidomide have distinct functional effects on erythropoiesis. Blood 118, 2296–2304 (2011)
Blackshear, P. J. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30, 945–952 (2002)
Stumpo, D. J. et al. Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood 114, 2401–2410 (2009)
Hudson, B. P., Martinez-Yamout, M. A., Dyson, H. J. & Wright, P. E. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nature Struct. Mol. Biol. 11, 257–264 (2004)
Schoenberg, D. R. & Maquat, L. E. Regulation of cytoplasmic mRNA decay. Nature Rev. Genet. 13, 246–259 (2012)
Novershtern, N. et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144, 296–309 (2011)
Brand, M. et al. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nature Struct. Mol. Biol. 11, 73–80 (2004)
Caterina, J. J., Donze, D., Sun, C. W., Ciavatta, D. J. & Townes, T. M. Cloning and functional characterization of LCR-F1: a bZIP transcription factor that activates erythroid-specific, human globin gene expression. Nucleic Acids Res. 22, 2383–2391 (1994)
Teittinen, K. J. et al. SAP30L (Sin3A-associated protein 30-like) is involved in regulation of cardiac development and hematopoiesis in zebrafish embryos. J. Cell. Biochem. 113, 3843–3852 (2012)
Shi, Z. T. et al. Protein 4.1R-deficient mice are viable but have erythroid membrane skeleton abnormalities. J. Clin. Invest. 103, 331–340 (1999)
Wang, Q., Khillan, J., Gadue, P. & Nishikura, K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290, 1765–1768 (2000)
Tibbetts, A. S. & Appling, D. R. Compartmentalization of mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 30, 57–81 (2010)
Kumkhaek, C. et al. MASL1 induces erythroid differentiation in human erythropoietin-dependent CD34+ cells through the Raf/MEK/ERK pathway. Blood 121, 3216–3227 (2013)
Zhang, L., Flygare, J., Wong, P., Lim, B. & Lodish, H. F. miR-191 regulates mouse erythroblast enucleation by down-regulating Riok3 and Mxi1. Genes Dev. 25, 119–124 (2011)
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008)
Acknowledgements
This work is supported by NIH grant P01 HL 32262 to H.F.L.; L.Z. is supported by a graduate fellowship from Singapore-Massachusetts Institute of Technology Alliance. We thank C. Sieff, J. Zhang, and P. Ji for providing reagents, T. Chavarria and F. Reinhardt for assisting with mouse transplantation experiments, J. Shih for helping with the making of constructs, M. Bousquet and C. Patterson for discussion and assistance with mouse transplantation experiment and antibody testing, S. Gupta, J. Kwon and I. Barrasa for processing raw RNA-seq data, processing microarray and discussion of bioinformatics analyses, and Sanofi Aventis for providing many GR agonists.
Author information
Authors and Affiliations
Contributions
L.Z. conceived the project, designed and performed the experiments and bioinformatics analysis, analysed the data, and wrote the paper. L.P. assisted with luciferase reporter assays and BFU–E isolation. V.R.-E. performed the GR Chip-seq experiment. P.T. provided training in bioinformatics analyses. J.F. performed the initial experiments with the GR partial agonists. B.L. supervised part of the research. H.F.L. supervised the research and edited the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-17, Supplementary Table 4 and an additional reference. (PDF 1589 kb)
Supplementary Table 1
This file contains Supplementary Table 1 with legend, it describes Zfp36l2 as the most abundant gene that is upregulated by all GR agonists that stimulate BFU-E self- renewal. BFU-Es were cultured in self-renewal medium with indicated GR agonists for 4 hours and the gene expression pattern was profiled by RNA-seq. The expression levels of several genes that are commonly induced by functional GR agonists are shown as RPKM values. (XLSX 11 kb)
Supplementary Table 2
This file contains Supplementary Table 2 with its legend, listing mRNAs preferentially bound to Zfp36l2 in BFU-E cells. RIP-chip was performed on freshly isolated BFU-Es, where Zfp36l2 antibody or control IgG was incubated with BFU-E cell lysis and microarrays were performed on immunoprecipitated mRNAs. The microarray probes with relative intensity, calculated as a ratio between its intensity in the Zfp36l2 immunoprecipitates relative to control IgG immunoprecipitates. Those with a ratio of greater than 1.5 in both biological repeats are listed. (XLSX 245 kb)
Supplementary Table 3
This file contains Supplementary Table 3 with its legend, listing Zfp36l2 functional target genes. Microarray gene expression profiling was carried out on day 3 of in vitro cultured BFU-E infected with control shRNA or Zfp36l2 shRNAs. A group of genes were identified with lower expression levels in BFU-Es cultured with DEX, compared with their counterparts in BFU-Es cultured without DEX. The repression of several of these DEX repressed genes is mediated by Zfp36l2, as knockdown of Zfp36l2 eliminates this effect. The potential Zfp36l2 functional target genes were then identified by intersecting these repressed genes with the group of genes identified by RIP-chip assay. (XLSX 11 kb)
Rights and permissions
About this article
Cite this article
Zhang, L., Prak, L., Rayon-Estrada, V. et al. ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors. Nature 499, 92–96 (2013). https://doi.org/10.1038/nature12215
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12215
This article is cited by
-
Associations of obesity and body shape with erythrocyte and reticulocyte parameters in the UK Biobank cohort
BMC Endocrine Disorders (2023)
-
Pre-configuring chromatin architecture with histone modifications guides hematopoietic stem cell formation in mouse embryos
Nature Communications (2022)
-
Formaldehyde-induced hematopoietic stem and progenitor cell toxicity in mouse lung and nose
Archives of Toxicology (2021)
-
Transfer learning efficiently maps bone marrow cell types from mouse to human using single-cell RNA sequencing
Communications Biology (2020)
-
Nuclear interacting SET domain protein 1 inactivation impairs GATA1-regulated erythroid differentiation and causes erythroleukemia
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.