Abstract
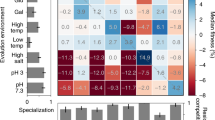
It is not known whether natural selection can encounter any given phenotype that can be produced by genetic variation. There has been a long-lasting debate about the processes that limit adaptation1,2,3,4,5,6 and, consequently, about how well adapted phenotypes are. Here we examine how development may affect adaptation, by decomposing the genotype–fitness map—the association between each genotype and its fitness—into two: one mapping genotype to phenotype by means of a computational model of organ development7, and one mapping phenotype to fitness. In the map of phenotype and fitness, the fitness of each individual is based on the similarity between realized morphology and optimal morphology. We use three different simulations to map phenotype to fitness, and these differ in the way in which similarity is calculated: similarity is calculated for each trait (in terms of each cell position individually), for a large or a small number of phenotypic landmarks (the ‘many-traits’ and ‘few-traits’ phenotype–fitness maps), and by measuring the overall surface roughness of morphology (the ‘roughness’ phenotype–fitness map). Evolution is simulated by applying the genotype–phenotype map and one phenotype–fitness map to each individual in the population, as well as random mutation and drift. We show that the complexity of the genotype–phenotype map prevents substantial adaptation in some of the phenotype–fitness maps: sustained adaptation is only possible using ‘roughness’ or ‘few-traits’ phenotype–fitness maps. The results contribute developmental understanding to the long-standing question of which aspects of phenotype can be effectively optimized by natural selection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Haldane, J. B. S. The Causes of Evolution (, Harper Brothers, 1932)
Wright, S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. Proc. VI Intern. Congress Genet. 1, 356–366 (1932)
Gould, S. J. & Lewontin, R. C. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist program. Proc. R. Soc. Lond. B 205, 581–598 (1979)
Alberch, P. In Evolution and Development Dahlem Konferenzen (ed. Bonner, J. T.) 313–332 (Springer, 1982)
Lande, R. & Arnold, S. J. The measurement of selection on correlated characters. Evolution 37, 1210–1226 (1983)
Fontana, W. Modelling 'evo-devo' with RNA. Bioessays 24, 1164–1177 (2002)
Salazar-Ciudad, I. & Jernvall, J. A computational model of teeth and the developmental origins of morphological variation. Nature 464, 583–586 (2010)
Waddington, C. H. The Strategy of the Genes (George Allen and Unwin, 1957)
Huynen, M. A., Stadler, P. F. & Fontana, W. Smoothness within ruggedness: the role of neutrality in adaptation. Proc. Natl Acad. Sci. USA 93, 397–401 (1996)
Ferrada, E. & Wagner, A. A comparison of genotype-–phenotype maps for RNA and proteins. Biophys. J. 102, 1916–1925 (2012)
Salazar-Ciudad, I. Developmental constraints vs. variational properties: how pattern formation can help to understand evolution and development. J. Exp. Zoolog. B 306B, 107–125 (2006)
Kauffman, S. A. The Origins of Order (Oxford Univ. Press, 1993)
Wagner, A. Evolution of gene networks by gene duplications: a mathematical model and its implications on genome organization. Proc. Natl Acad. Sci. USA 91, 4387–4391 (1994)
Hansen, T. F. & Wagner, G. P. Modeling genetic architecture: a multilinear theory of gene interaction. Theor. Popul. Biol. 59, 61–86 (2001)
Salazar-Ciudad, I. & Jernvall, J. How different types of pattern formation mechanisms affect the evolution of form and development. Evol. Dev. 6, 6–16 (2004)
Evans, A. R. et al. High-level similarity of dentitions in carnivorans and rodents. Nature 445, 78–81 (2007)
Orzack, S. H. & Sober, E. Optimality models and the test of adaptationism. Am. Nat. 143, 361–380 (1994)
Maynard Smith, J. Optimization theory in evolution. Ann. Rev. Ecol. Syst. 9, 31–56 (1978)
Ji, Q. et al. The earliest known eutherian mammal. Nature 416, 816–822 (2002)
Charles, C. et al. Regulation of tooth number by fine-tuning levels of receptor-tyrosine kinase signaling. Development 138, 4063–4073 (2011)
Klingenberg, C. P. Morphometrics and the role of the phenotype in studies of the evolution of developmental mechanisms. Gene 287, 3–10 (2002)
Bunn, J. M. et al. Comparing Dirichlet normal surface energy of tooth crowns, a new technique of molar shape quantification for dietary inference, with previous methods in isolation and in combination. Am. J. Phys. Anthropol. 145, 247–261 (2011)
Alfaro, M. E. et al. Evolutionary consequences of many-to-one mapping of jaw morphology to mechanics in labrid fishes. Am. Nat. 165, E140–E154 (2005)
Santana, S. E., Strait, S. & Dumont, E. R. The better to eat you with: functional correlates of tooth structure in bats. Funct. Ecol. 25, 839–847 (2011)
Godfrey, L. R. et al. Dental topography indicates ecological contraction of lemur communities. Am. J. Phys. Anthropol. 148, 215–227 (2012)
Charlesworth, B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nature Rev. Genet. 10, 195–205 (2009)
Tenaillon, O. et al. Quantifying organismal complexity using a population genetic approach. PLoS ONE 2, e217 (2007)
Polly, P. D., Le Comber, S. C. & Burland, T. M. On the occlusal fit of tribosphenic molars: Are we underestimating species diversity in the Mesozoic? J. Mamm. Evol. 12, 283–299 (2005)
Orr, H. A. The population genetics of adaptation on correlated fitness landscapes: the block model. Evolution 60, 1113–1124 (2006)
Acknowledgements
We thank J. Jernvall, M. Brun Usan, I. Salvador Martinez, A. Matamoro, S. Newman and R. Zimm for comments and the CSC (IT Center for Science). This research was funded by the Finnish Academy (WBS 1250271) and the Spanish Ministry of Science and Innovation (BFU2010-17044).
Author information
Authors and Affiliations
Contributions
I.S.-C. conceived the study; and M.M.-R and I.S.-C. constructed the evolutionary model. M.M.-R carried out computer simulations and quantitative analyses. I.S.-C and M.M.-R. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-14, Supplementary Methods and Supplementary References. (PDF 5792 kb)
Rights and permissions
About this article
Cite this article
Salazar-Ciudad, I., Marín-Riera, M. Adaptive dynamics under development-based genotype–phenotype maps. Nature 497, 361–364 (2013). https://doi.org/10.1038/nature12142
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12142
This article is cited by
-
Using phenotypic plasticity to understand the structure and evolution of the genotype–phenotype map
Genetica (2022)
-
Development and selective grain make plasticity 'take the lead' in adaptive evolution
BMC Ecology and Evolution (2021)
-
Why call it developmental bias when it is just development?
Biology Direct (2021)
-
Phylogenetic and Developmental Constraints Dictate the Number of Cusps on Molars in Rodents
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.