Abstract
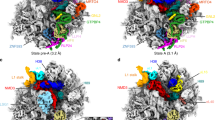
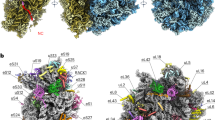
Protein synthesis in all cells is carried out by macromolecular machines called ribosomes. Although the structures of prokaryotic, yeast and protist ribosomes have been determined, the more complex molecular architecture of metazoan 80S ribosomes has so far remained elusive. Here we present structures of Drosophila melanogaster and Homo sapiens 80S ribosomes in complex with the translation factor eEF2, E-site transfer RNA and Stm1-like proteins, based on high-resolution cryo-electron-microscopy density maps. These structures not only illustrate the co-evolution of metazoan-specific ribosomal RNA with ribosomal proteins but also reveal the presence of two additional structural layers in metazoan ribosomes, a well-ordered inner layer covered by a flexible RNA outer layer. The human and Drosophila ribosome structures will provide the basis for more detailed structural, biochemical and genetic experiments.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Accessions
Protein Data Bank
Data deposits
Coordinates of the atomic models have been deposited in the Protein Data Bank with accession numbers 3J38, 3J39, 3J3C and 3J3E for Drosophila 80S ribosomes and 3J3A, 3J3B, 3J3D and 3J3F for human 80S ribosomes. Full models can be obtained from the database of aligned ribosomal complexes (DARC) site (http://darcsite.genzentrum.lmu.de/darc/). Electron-microscopy maps of the Drosophila and human ribosomes have been deposited in the EM Data Bank under the accession codes EMD-5591 and EMD-5592, respectively. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper. Correspondence and requests for materials should be addressed to R.B. (beckmann@lmb.uni-muenchen.de).
References
Schmeing, T. M. & Ramakrishnan, V. What recent ribosome structures have revealed about the mechanism of translation. Nature 461, 1234–1242 (2009)
Wilson, D. N. & Cate, J. H. D. The structure and function of the eukaryotic ribosome. Cold Spring Harb. Perspect. Biol. 4, a011536 (2012)
Klinge, S., Voigts-Hoffmann, F., Leibundgut, M. & Ban, N. Atomic structures of the eukaryotic ribosome. Trends Biochem. Sci. 37, 189–198 (2012)
Melnikov, S. et al. One core, two shells: bacterial and eukaryotic ribosomes. Nature Struct. Mol. Biol. 19, 560–567 (2012)
Taylor, D. J. et al. Comprehensive molecular structure of the eukaryotic ribosome. Structure 17, 1591–1604 (2009)
Armache, J. P. et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl Acad. Sci. USA 107, 19748–19753 (2010)
Armache, J. P. et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-A cryo-EM map of the 80S eukaryotic ribosome. Proc. Natl Acad. Sci. USA 107, 19754–19759 (2010)
Rabl, J., Leibundgut, M., Ataide, S. F., Haag, A. & Ban, N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331, 730–736 (2011)
Klinge, S., Voigts-Hoffmann, F., Leibundgut, M., Arpagaus, S. & Ban, N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334, 941–948 (2011)
Ben-Shem, A. et al. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334, 1524–1529 (2011)
Dube, P. et al. Correlation of the expansion segments in mammalian rRNA with the fine structure of the 80 S ribosome; a cryoelectron microscopic reconstruction of the rabbit reticulocyte ribosome at 21 A resolution. J. Mol. Biol. 279, 403–421 (1998)
Spahn, C. M. et al. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes; the IRES functions as an RNA-based translation factor. Cell 118, 465–475 (2004)
Boehringer, D., Thermann, R., Ostareck-Lederer, A., Lewis, J. D. & Stark, H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure 13, 1695–1706 (2005)
Chandramouli, P. et al. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure 16, 535–548 (2008)
Ruvinsky, I. & Meyuhas, O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem. Sci. 31, 342–348 (2006)
Koyama, Y., Katagiri, S., Hanai, S., Uchida, K. & Miwa, M. Poly(ADP-ribose) polymerase interacts with novel Drosophila ribosomal proteins, L22 and l23a, with unique histone-like amino-terminal extensions. Gene 226, 339–345 (1999)
Ramakrishnan, V. Histone structure and the organization of the nucleosome. Annu. Rev. Biophys. Biomol. Struct. 26, 83–112 (1997)
Taylor, D. J. et al. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 26, 2421–2431 (2007)
Harms, J. M. et al. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell 30, 26–38 (2008)
Gao, Y. G. et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694–699 (2009)
Dever, T. E. & Green, R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 4, a013706 (2012)
Ogle, J. M. & Ramakrishnan, V. Structural insights into translational fidelity. Annu. Rev. Biochem. 74, 129–177 (2005)
Demeshkina, N., Jenner, L., Westhof, E., Yusupov, M. & Yusupova, G. A new understanding of the decoding principle on the ribosome. Nature 484, 256–259 (2012)
Lu, H., Li, W., Noble, W. S., Payan, D. & Anderson, D. C. Riboproteomics of the hepatitis C virus internal ribosomal entry site. J. Proteome Res. 3, 949–957 (2004)
Spahn, C. M. et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291, 1959–1962 (2001)
Balagopal, V. & Parker, R. Stm1 modulates translation after 80S formation in Saccharomyces cerevisiae. RNA 17, 835–842 (2011)
Gerbi, S. A. in Ribosomal RNA—Structure, Evolution, Processing, and Function in Protein Synthesis (eds Zimmermann, R. A. & Dahlberg, A. E.) 71–87 (CRC Press, 1996)
Haga, J. Y., Hamilton, M. G. & Petermann, M. L. Electron microscopic observations on the large subunit of the rat liver ribosome. J. Cell Biol. 47, 211–221 (1970)
Cannone, J. J. et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3, 2 (2002)
Fields, D. S. & Gutell, R. R. An analysis of large rRNA sequences folded by a thermodynamic method. Fold. Des. 1, 419–430 (1996)
Alkemar, G. & Nygard, O. Probing the secondary structure of expansion segment ES6 in 18S ribosomal RNA. Biochemistry 45, 8067–8078 (2006)
Andersen, C. B. et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature 443, 663–668 (2006)
Srivastava, S., Verschoor, A. & Frank, J. Eukaryotic initiation factor-3 does not prevent association through physical blockage of the ribosomal subunit-subunit interface. J. Mol. Biol. 226, 301–304 (1992)
Siridechadilok, B., Fraser, C. S., Hall, R. J., Doudna, J. A. & Nogales, E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310, 1513–1515 (2005)
Yu, Y., Abaeva, I. S., Marintchev, A., Pestova, T. V. & Hellen, C. U. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 39, 4851–4865 (2011)
Beckmann, R. et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107, 361–372 (2001)
Becker, T. et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326, 1369–1373 (2009)
Sweeney, R., Chen, L. H. & Yao, M. C. An rRNA variable region has an evolutionarily conserved essential role despite sequence divergence. Mol. Cell. Biol. 14, 4203–4215 (1994)
Bradatsch, B. et al. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nature Struct. Mol. Biol. 19, 1234–1241 (2012)
Greber, B. J., Boehringer, D., Montellese, C. & Ban, N. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nature Struct. Mol. Biol. 19, 1228–1233 (2012)
Leidig, C. et al. Structural characterization of a eukaryotic chaperone—the ribosome-associated complex. Nature Struct. Mol. Biol. 20, 23–28 (2013)
Blau, M. et al. ERj1p uses a universal ribosomal adaptor site to coordinate the 80S ribosome at the membrane. Nature Struct. Mol. Biol. 12, 1015–1016 (2005)
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996)
Jossinet, F. & Westhof, E. Sequence to Structure (S2S): display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics 21, 3320–3321 (2005)
Jossinet, F., Ludwig, T. E. & Westhof, E. Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics 26, 2057–2059 (2010)
Eswar, N., Eramian, D., Webb, B., Shen, M. Y. & Sali, A. Protein structure modeling with MODELLER. Methods Mol. Biol. 426, 145–159 (2008)
Jenner, L., Demeshkina, N., Yusupova, G. & Yusupov, M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nature Struct. Mol. Biol. 17, 1072–1078 (2010)
Gebauer, F., Corona, D. F., Preiss, T., Becker, P. B. & Hentze, M. W. Translational control of dosage compensation in Drosophila by Sex-lethal: cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J. 18, 6146–6154 (1999)
Fuss, I. J., Kanof, M. E., Smith, P. D. & Zola, H. Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr. Protoc. Immunol. 85, 7.1.1–7.1.8 (2009)
Becker, T. et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482, 501–506 (2012)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Becker, T. et al. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nature Struct. Mol. Biol. 18, 715–720 (2011)
Hirsch, M., Scholkopf, B. & Habeck, M. A blind deconvolution approach for improving the resolution of cryo-EM density maps. J. Comput. Biol. 18, 335–346 (2011)
Lasker, K. et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl Acad. Sci. USA 109, 1380–1387 (2012)
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001)
Maden, B. E. et al. Clones of human ribosomal DNA containing the complete 18 S-rRNA and 28 S-rRNA genes. Characterization, a detailed map of the human ribosomal transcription unit and diversity among clones. Biochem. J. 246, 519–527 (1987)
Tautz, D., Hancock, J. M., Webb, D. A., Tautz, C. & Dover, G. A. Complete sequences of the rRNA genes of Drosophila melanogaster. Mol. Biol. Evol. 5, 366–376 (1988)
Thompson, J. F., Wegnez, M. R. & Hearst, J. E. Determination of the secondary structure of Drosophila melanogaster 5 S RNA by hydroxymethyltrimethylpsoralen crosslinking. J. Mol. Biol. 147, 417–436 (1981)
Rousset, F., Pelandakis, M. & Solignac, M. Evolution of compensatory substitutions through G.U intermediate state in Drosophila rRNA. Proc. Natl Acad. Sci. USA 88, 10032–10036 (1991)
Dunkle, J. A. et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332, 981–984 (2011)
Hofacker, I. L. Vienna RNA secondary structure server. Nucleic Acids Res. 31, 3429–3431 (2003)
Trabuco, L. G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008)
Humphrey, W., Dalke, A. & Schulten, K. VMD - Visual Molecular Dynamics. J. Mol. Graph. 14, 33–38 (1996)
Emsley, P. & Cowtan, K. Coot: model-Building Tools for Molecular Graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004)
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011)
Katoh, K., Kuma, K., Toh, H. & Miyata, T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (2005)
Pettersen, E. F. et al. UCSF Chimera — a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Nierhaus, K. H. & Dohme, F. Total reconstitution of functionally active 50S ribosomal subunits from E. coli. Proc. Natl Acad. Sci. USA 71, 4713–4717 (1974)
Márquez, V. et al. Proteomic characterization of archaeal ribosomes reveals the presence of novel archaeal-specific ribosomal proteins. J. Mol. Biol. 405, 1215–1232 (2011)
Norousi, R. et al. Automated post-picking using MAPPOS improves particle image detection from cryo-EM micrographs. J. Struct. Biol. http://dx.doi.org/10.1016/j.jsb.2013.02.008 (2013)
Acknowledgements
We thank C. Ungewickell for assistance with cryo-EM data collection and P. Palluch for preparation of peripheral blood mononucleic cells. We thank M. Yusupov, A. Ben-Shem, N. Garreau de Loubresse and S. Melnikov for sharing S. cerevisiae X-ray data before publication. We thank P. Becker for access to his fly facility and help with embryo collection, and V. Márquez, T. Fröhlich, G. Arnold, I. Forné and A. Imhof for mass-spectrometry analysis. This research was supported by grants from the Deutsche Forschungsgemeinschaft SFB594, SFB646 and GRK 1721 (to R.B.), and FOR1805 (to R.B. and D.N.W.). D.N.W. is supported by the European Molecular Biology Organization (EMBO) young investigator program. This work was supported by a European Research Council (ERC) Advanced Grant (to R.B.).
Author information
Authors and Affiliations
Contributions
A.M.A. prepared D. melanogaster embryo extracts, purified D. melanogaster and H. sapiens ribosome samples, carried out mass-spectrometry analysis of H. sapiens ribosomes and prepared the figures; A.M.A. and J.-P.A. contributed blood, processed cryo-EM data and built atomic models; O.B. carried out cryo-EM data collection; M.H. performed deconvolution and sharpening on electron density maps; M.S. designed experiments for blood collection and peripheral-blood-mononuclear-cell preparations for human ribosome purification; A.M.A., J.-P.A., D.N.W. and R.B. interpreted results and wrote the manuscript. D.N.W. and R.B. designed research and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-24, and Supplementary Tables 1-4. (PDF 11725 kb)
Rights and permissions
About this article
Cite this article
Anger, A., Armache, JP., Berninghausen, O. et al. Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85 (2013). https://doi.org/10.1038/nature12104
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12104
This article is cited by
-
A molecular network of conserved factors keeps ribosomes dormant in the egg
Nature (2023)
-
In vivo secondary structural analysis of Influenza A virus genomic RNA
Cellular and Molecular Life Sciences (2023)
-
A multi-targeting drug design strategy for identifying potent anti-SARS-CoV-2 inhibitors
Acta Pharmacologica Sinica (2022)
-
A stem cell roadmap of ribosome heterogeneity reveals a function for RPL10A in mesoderm production
Nature Communications (2022)
-
Chemical reversible crosslinking enables measurement of RNA 3D distances and alternative conformations in cells
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.