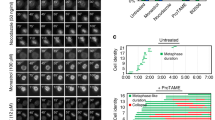
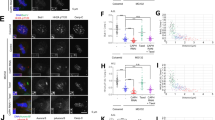
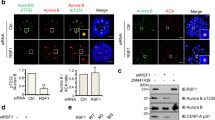
Abstract
Accurate segregation of the replicated genome requires chromosome biorientation on the spindle. Biorientation is ensured by Aurora B kinase (Ipl1), a member of the four-subunit chromosomal passenger complex (CPC)1,2. Localization of the CPC to the inner centromere is central to the current model for how tension ensures chromosome biorientation: kinetochore–spindle attachments that are not under tension remain close to the inner centromere and are destabilized by Aurora B phosphorylation, whereas kinetochores under tension are pulled away from the influence of Aurora B, stabilizing their microtubule attachments3,4,5. Here we show that an engineered truncation of the Sli15 (known as INCENP in humans) subunit of budding yeast CPC that eliminates association with the inner centromere nevertheless supports proper chromosome segregation during both mitosis and meiosis. Truncated Sli15 suppresses the deletion phenotypes of the inner-centromere-targeting proteins survivin (Bir1), borealin (Nbl1), Bub1 and Sgo1 (ref. 6). Unlike wild-type Sli15, truncated Sli15 localizes to pre-anaphase spindle microtubules. Premature targeting of full-length Sli15 to microtubules by preventing Cdk1 (also known as Cdc28) phosphorylation also suppresses the inviability of Bir1 deletion. These results suggest that activation of Aurora B kinase by clustering either on chromatin or on microtubules is sufficient for chromosome biorientation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ruchaud, S., Carmena, M. & Earnshaw, W. C. Chromosomal passengers: conducting cell division. Nature Rev. Mol. Cell Biol. 8, 798–812 (2007)
Pinsky, B. A. & Biggins, S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 15, 486–493 (2005)
Tanaka, T. U. et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108, 317–329 (2002)
Liu, D., Vader, G., Vromans, M. J. M., Lampson, M. A. & Lens, S. M. A. Sensing chromosome bi-orientation by spatial separation of Aurora B kinase from kinetochore substrates. Science 323, 1350–1353 (2009)
Lampson, M. A. & Cheeseman, I. M. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21, 133–140 (2011)
Kawashima, S. A., Yamagishi, Y., Honda, T., Ishiguro, K.-I. & Watanabe, Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327, 172–177 (2010)
Yoon, H. J. & Carbon, J. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc. Natl Acad. Sci. USA 96, 13208–13213 (1999)
Cho, U.-S. & Harrison, S. C. Ndc10 is a platform for inner kinetochore assembly in budding yeast. Nature Struct. Mol. Biol. 19, 48–55 (2012)
Yamagishi, Y., Honda, T., Tanno, Y. & Watanabe, Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science 330, 239–243 (2010)
Kelly, A. E. et al. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330, 235–239 (2010)
Wang, F. et al. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 330, 231–235 (2010)
Shimogawa, M. M., Widlund, P. O., Riffle, M., Ess, M. & Davis, T. N. Bir1 is required for the tension checkpoint. Mol. Biol. Cell 20, 915–923 (2009)
Makrantoni, V. & Stark, M. Efficient chromosome bi-orientation and the tension checkpoint in Saccharomyces cerevisiae both require Bir1. Mol. Cell. Biol. 29, 4552–4562 (2009)
Sandall, S. et al. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell 127, 1179–1191 (2006)
Jeyaprakash, A. A. et al. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 131, 271–285 (2007)
Biggins, S. & Murray, A. W. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15, 3118–3129 (2001)
Kim, J. H., Kang, J. S. & Chan, C. S. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 145, 1381–1394 (1999)
Indjeian, V. B. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science 307, 130–133 (2005)
Li, R. & Murray, A. W. Feedback control of mitosis in budding yeast. Cell 66, 519–531 (1991)
Ng, T. M., Waples, W. G., Lavoie, B. D. & Biggins, S. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol. Biol. Cell 20, 3818–3827 (2009)
Shonn, M. A., McCarroll, R. & Murray, A. W. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 289, 300–303 (2000)
Pereira, G. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 302, 2120–2124 (2003)
Rozelle, D. K., Hansen, S. D. & Kaplan, K. B. Chromosome passenger complexes control anaphase duration and spindle elongation via a kinesin-5 brake. J. Cell Biol. 193, 285–294 (2011)
Nakajima, Y. et al. Ipl1/Aurora-dependent phosphorylation of Sli15/INCENP regulates CPC–spindle interaction to ensure proper microtubule dynamics. J. Cell Biol. 194, 137–153 (2011)
Sessa, F. et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell 18, 379–391 (2005)
Kelly, A. E. et al. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev. Cell 12, 31–43 (2007)
Tseng, B. S., Tan, L., Kapoor, T. M. & Funabiki, H. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev. Cell 18, 903–912 (2010)
Wan, X. et al. Protein architecture of the human kinetochore microtubule attachment site. Cell 137, 672–684 (2009)
Longtine, M. S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998)
Hieter, P., Mann, C., Snyder, M. & Davis, R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell 40, 381–392 (1985)
Acknowledgements
The authors would like to thank the Desai and Oegema laboratories for discussions; S. Sandall, H. Hu and E. Manrriquez for assistance; B. Ren’s laboratory for help with ChIP experiments; S. Biggins, D. Dawson, G. Pereira, G. Barnes, P. Hieter and the Yeast Resource Center for strains and plasmids; and K. Oegema, R. Green and J. DeLuca for comments on the manuscript. This work was supported by a National Institutes of Health (NIH) grant (GM074215) to A.D. and a Damon Runyon Cancer Research Foundation Fellowship (DRG 2007-09) to C.S.C. A.D. receives salary and other support from the Ludwig Institute for Cancer Research.
Author information
Authors and Affiliations
Contributions
C.S.C. and A.D. designed experiments and wrote the manuscript. C.S.C. performed the experiments and analysed the data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-3, Supplementary Table 1, a Supplementary Discussion and Supplementary References. (PDF 1457 kb)
Rights and permissions
About this article
Cite this article
Campbell, C., Desai, A. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature 497, 118–121 (2013). https://doi.org/10.1038/nature12057
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12057
This article is cited by
-
Tension can directly suppress Aurora B kinase-triggered release of kinetochore-microtubule attachments
Nature Communications (2022)
-
Killing two birds with one stone: how budding yeast Mps1 controls chromosome segregation and spindle assembly checkpoint through phosphorylation of a single kinetochore protein
Current Genetics (2020)
-
The binding of Borealin to microtubules underlies a tension independent kinetochore-microtubule error correction pathway
Nature Communications (2019)
-
Complete microtubule–kinetochore occupancy favours the segregation of merotelic attachments
Nature Communications (2018)
-
Inner centromere localization of the CPC maintains centromere cohesion and allows mitotic checkpoint silencing
Nature Communications (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.