Abstract
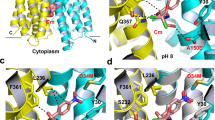
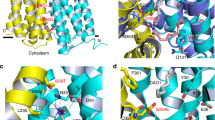
Multidrug and toxic compound extrusion (MATE) family transporters are conserved in the three primary domains of life (Archaea, Bacteria and Eukarya), and export xenobiotics using an electrochemical gradient of H+ or Na+ across the membrane1,2. MATE transporters confer multidrug resistance to bacterial pathogens3,4,5,6 and cancer cells7, thus causing critical reductions in the therapeutic efficacies of antibiotics and anti-cancer drugs, respectively. Therefore, the development of MATE inhibitors has long been awaited in the field of clinical medicine8,9. Here we present the crystal structures of the H+-driven MATE transporter from Pyrococcus furiosus in two distinct apo-form conformations, and in complexes with a derivative of the antibacterial drug norfloxacin and three in vitro selected thioether-macrocyclic peptides, at 2.1–3.0 Å resolutions. The structures, combined with functional analyses, show that the protonation of Asp 41 on the amino (N)-terminal lobe induces the bending of TM1, which in turn collapses the N-lobe cavity, thereby extruding the substrate drug to the extracellular space. Moreover, the macrocyclic peptides bind the central cleft in distinct manners, which correlate with their inhibitory activities. The strongest inhibitory peptide that occupies the N-lobe cavity may pave the way towards the development of efficient inhibitors against MATE transporters.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
The coordinates and structure factors for the P. furiosus apo MATE (‘straight’ and ‘bent’ conformations), P26A mutant, Br-NRF-bound MATE and peptide-bound MATEs have been deposited in the Protein Data Bank, under the accession numbers 3VVN, 3VVO, 3W4T, 3VVP, 3VVQ, 3VVR and 3VVS, respectively.
References
Brown, M. H., Paulsen, I. T. & Skurray, R. A. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 31, 394–395 (1999)
He, G. X. et al. An H+-coupled multidrug efflux pump, PmpM, a member of the MATE family of transporters, from Pseudomonas aeruginosa. J. Bacteriol. 186, 262–265 (2004)
Kaatz, G. W., McAleese, F. & Seo, S. M. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 49, 1857–1864 (2005)
McAleese, F. et al. A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to tigecycline. Antimicrob. Agents Chemother. 49, 1865–1871 (2005)
Tsuda, M. et al. Oppositely directed H+ gradient functions as a driving force of rat H+/organic cation antiporter MATE1. Am. J. Physiol. 292, F593–F598 (2007)
Becker, M. L. et al. Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: a preliminary study. Diabetes 58, 745–749 (2009)
Minematsu, T. & Giacomini, K. M. Interactions of tyrosine kinase inhibitors with organic cation transporters and multidrug and toxic compound extrusion proteins. Mol. Cancer Ther. 10, 531–539 (2011)
Ito, S. et al. Potent and specific inhibition of mMate1-mediated efflux of type I organic cations in the liver and kidney by pyrimethamine. J. Pharmacol. Exp. Ther. 333, 341–350 (2010)
Kusuhara, H. et al. Effects of a MATE protein inhibitor, pyrimethamine, on the renal elimination of metformin at oral microdose and at therapeutic dose in healthy subjects. Clin. Pharmacol. Ther. 89, 837–844 (2011)
Dawson, R. J. & Locher, K. P. Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 (2006)
Murakami, S., Nakashima, R., Yamashita, E., Matsumoto, T. & Yamaguchi, A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443, 173–179 (2006)
Wasaznik, A., Grinholc, M. & Bielawski, K. P. Active efflux as the multidrug resistance mechanism. Postepy Hig. Med. Dosw. 63, 123–133 (2009)
Morita, Y. et al. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42, 1778–1782 (1998)
Morita, Y., Kataoka, A., Shiota, S., Mizushima, T. & Tsuchiya, T. NorM of Vibrio parahaemolyticus is an Na+-driven multidrug efflux pump. J. Bacteriol. 182, 6694–6697 (2000)
Otsuka, M. et al. Identification of essential amino acid residues of the NorM Na+/multidrug antiporter in Vibrio parahaemolyticus. J. Bacteriol. 187, 1552–1558 (2005)
He, X. et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467, 991–994 (2010)
Law, C. J., Maloney, P. C. & Wang, D. N. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62, 289–305 (2008)
Cherezov, V. Lipidic cubic phase technologies for membrane protein structural studies. Curr. Opin. Struct. Biol. 21, 559–566 (2011)
Hipolito, C. J. & Suga, H. Ribosomal production and in vitro selection of natural product-like peptidomimetics: the FIT and RaPID systems. Curr. Opin. Chem. Biol. 16, 196–203 (2012)
Hayashi, Y., Morimoto, J. & Suga, H. In vitro selection of anti-Akt2 thioether-macrocyclic peptides leading to isoform-selective inhibitors. ACS Chem. Biol. 7, 607–613 (2012)
Yamagishi, Y. et al. Natural product-like macrocyclic N-methyl-peptide inhibitors against a ubiquitin ligase uncovered from a ribosome-expressed de novo library. Chem. Biol. 18, 1562–1570 (2011)
Morimoto, J., Hayashi, Y. & Suga, H. Discovery of macrocyclic peptides armed with a mechanism-based warhead: isoform-selective inhibition of human deacetylase SIRT2. Angew. Chem. Int. Ed. 51, 3423–3427 (2012)
Altschul, S. F. et al. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272, 5101–5109 (2005)
Koga, H., Itoh, A., Murayama, S., Suzue, S. & Irikura, T. Structure-activity relationships of antibacterial 6,7- and 7,8-disubstituted 1-alkyl-1,4-dihydro-4-oxoquinoline-3-carboxylic acids. J. Med. Chem. 23, 1358–1363 (1980)
Lu, M. et al. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc. Natl Acad. Sci. USA 110, 2099–2104 (2013)
Flot, D. et al. The ID23–2 structural biology microfocus beamline at the ESRF. J. Synchrotron Radiat. 17, 107–118 (2010)
Tsujimoto, K., Semadeni, M., Huflejt, M. & Packer, L. Intracellular pH of halobacteria can be determined by the fluorescent dye 2′, 7′-bis(carboxyethyl)-5(6)-carboxyfluorescein. Biochem. Biophys. Res. Commun. 155, 123–129 (1988)
Li, X. Z., Poole, K. & Nikaido, H. Contributions of MexAB-OprM and an EmrE homolog to intrinsic resistance of Pseudomonas aeruginosa to aminoglycosides and dyes. Antimicrob. Agents Chemother. 47, 27–33 (2003)
Hirata, K. et al. New micro-beam beamline at SPring-8, targeting at protein micro-crystallography. AIP Conf. Proc. 1234, 901–904 (2010)
Xu, H., Smith, A. B., Sahinidis, N. V. & Weeks, C. M. SnB version 2.3: triplet sieve phasing for centrosymmetric structures. J. Appl. Cryst. 41, 644–646 (2008)
Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2007)
Eswar, N. et al. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 2, 2.9.1–2.9.31 (2007)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D 68, 368–380 (2012)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Zwart, P. H. et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 (2008)
Ito, K., Ebihara, K., Uno, M. & Nakamura, Y. Conserved motifs in prokaryotic and eukaryotic polypeptide release factors: tRNA-protein mimicry hypothesis. Proc. Natl Acad. Sci. USA 93, 5443–5448 (1996)
Martinac, B., Buechner, M., Delcour, A. H., Adler, J. & Kung, C. Pressure-sensitive ion channel in Escherichia coli. Proc. Natl Acad. Sci. USA 84, 2297–2301 (1987)
Kuo, M. M., Saimi, Y., Kung, C. & Choe, S. Patch clamp and phenotypic analyses of a prokaryotic cyclic nucleotide-gated K+ channel using Escherichia coli as a host. J. Biol. Chem. 282, 24294–24301 (2007)
Acknowledgements
We are grateful to the beam-line staff at BL32XU of SPring-8 for assistance in data collection (proposals 2011B1062, 2012A1087 and 2012B1161), and the RIKEN BioResource Center (Ibaraki, Japan) for providing the P. furiosus genomic DNA. We are also grateful to N. Dohmae and Y. Sugita (RIKEN Advanced Science Institute, Japan) for discussions. This work was supported by the Japan Society for the Promotion of Science (JSPS) through its ‘Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST program)’ to O.N.; by the Core Research for Evolutional Science and Technology Program ‘The Creation of Basic Medical Technologies to Clarify and Control the Mechanisms Underlying Chronic Inflammation’ of Japan Science and Technology Agency to O.N.; and by a Grant-in-Aid for Scientific Research (S) (24227004) and a Grant-in-Aid for Young Scientists (A) (22687007) from MEXT to O.N. and R.I., respectively. This work was also supported by a JSPS Grant-in-Aid for the Specially Promoted Research (21000005) and MEXT Platform for Drug Discovery, Informatics, and Structural Life Science to H.S., and a Grant-in-Aid for JSPS post-doctoral fellows to C.J.H. (P11344).
Author information
Authors and Affiliations
Contributions
Y.T. expressed and purified PfMATE for crystallization, collected the diffraction data, solved the structures and made the mutants for functional analyses. C.J.H. performed selections, syntheses and inhibition assays of macrocyclic peptides. A.D.M. performed the fluorescence analysis. K.I. performed growth complementation tests. T.K. performed drug susceptibility tests. T.H. synthesized Br-NRF. K.K. and H.E.K. assisted with data collection. M.H. and T.T. contributed to the early stage of the project. Y.T., C.J.H., H.E.K., R.I. and O.N. wrote the manuscript. H.S. and O.N. directed and supervised all of the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Supplementary Tables 1-2, Supplementary Figures 1-10 and Supplementary References. (PDF 5096 kb)
Proton-dependent drug extrusion by MATE
The structural transition between the “straight” and “bent” conformations, viewed from the extracellular and membrane sides. (MP4 3137 kb)
Rights and permissions
About this article
Cite this article
Tanaka, Y., Hipolito, C., Maturana, A. et al. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature 496, 247–251 (2013). https://doi.org/10.1038/nature12014
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12014
This article is cited by
-
Dynamics of efflux pumps in antimicrobial resistance, persistence, and community living of Vibrionaceae
Archives of Microbiology (2024)
-
Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn2+ uptake into the Golgi apparatus
Nature Communications (2023)
-
Engineered MATE multidrug transporters reveal two functionally distinct ion-coupling pathways in NorM from Vibrio cholerae
Communications Biology (2021)
-
Conserved functions of MATE transporters and their potential to enhance plant tolerance to aluminium toxicity
Journal of Plant Biochemistry and Biotechnology (2021)
-
A rice QTL GS3.1 regulates grain size through metabolic-flux distribution between flavonoid and lignin metabolons without affecting stress tolerance
Communications Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.