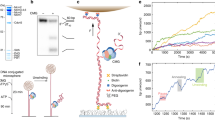
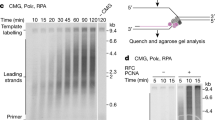
Abstract
Chromosomal replication machines contain coupled DNA polymerases that simultaneously replicate the leading and lagging strands1. However, coupled replication presents a largely unrecognized topological problem. Because DNA polymerase must travel a helical path during synthesis, the physical connection between leading- and lagging-strand polymerases causes the daughter strands to entwine, or produces extensive build-up of negative supercoils in the newly synthesized DNA2,3,4. How DNA polymerases maintain their connection during coupled replication despite these topological challenges is unknown. Here we examine the dynamics of the Escherichia coli replisome, using ensemble and single-molecule methods, and show that the replisome may solve the topological problem independent of topoisomerases. We find that the lagging-strand polymerase frequently releases from an Okazaki fragment before completion, leaving single-strand gaps behind. Dissociation of the polymerase does not result in loss from the replisome because of its contact with the leading-strand polymerase. This behaviour, referred to as ‘signal release’, had been thought to require a protein, possibly primase, to pry polymerase from incompletely extended DNA fragments5,6,7. However, we observe that signal release is independent of primase and does not seem to require a protein trigger at all. Instead, the lagging-strand polymerase is simply less processive in the context of a replisome. Interestingly, when the lagging-strand polymerase is supplied with primed DNA in trans, uncoupling it from the fork, high processivity is restored. Hence, we propose that coupled polymerases introduce topological changes, possibly by accumulation of superhelical tension in the newly synthesized DNA, that cause lower processivity and transient lagging-strand polymerase dissociation from DNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kornberg, A. & Baker, T. A. DNA Replication 2nd edn (W. H. Freeman, 1992)
Ullsperger, C. J., Volgodskii, A. V. & Cozzarelli, N. R. Unlinking of DNA by Topoisomerases during DNA Replication Vol. 9 (Springer, 1995)
Hingorani, M. M. & O’Donnell, M. Sliding clamps: a (tail)ored fit. Curr. Biol. 10, R25–R29 (2000)
Wyman, C. & Botchan, M. DNA replication. A familiar ring to DNA polymerase processivity. Curr. Biol. 5, 334–337 (1995)
Hamdan, S. M., Loparo, J. J., Takahashi, M., Richardson, C. C. & van Oijen, A. M. Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis. Nature 457, 336–339 (2009)
Li, X. & Marians, K. J. Two distinct triggers for cycling of the lagging strand polymerase at the replication fork. J. Biol. Chem. 275, 34757–34765 (2000)
Yang, J., Nelson, S. W. & Benkovic, S. J. The control mechanism for lagging strand polymerase recycling during bacteriophage T4 DNA replication. Mol. Cell 21, 153–164 (2006)
Johnson, A. & O’Donnell, M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 74, 283–315 (2005)
Postow, L., Peter, B. J. & Cozzarelli, N. R. Knot what we thought before: the twisted story of replication. Bioessays 21, 805–808 (1999)
Chastain, P. D., II, Makhov, A. M., Nossal, N. G. & Griffith, J. Architecture of the replication complex and DNA loops at the fork generated by the bacteriophage t4 proteins. J. Biol. Chem. 278, 21276–21285 (2003)
Vologodskii, A. V., Lukashin, A. V., Anshelevich, V. V. & Frank-Kamenetskii, M. D. Fluctuations in superhelical DNA. Nucleic Acids Res. 6, 967–982 (1979)
Leu, F. P., Georgescu, R. & O’Donnell, M. Mechanism of the E. coli tau processivity switch during lagging-strand synthesis. Mol. Cell 11, 315–327 (2003)
Espeli, O. & Marians, K. J. Untangling intracellular DNA topology. Mol. Microbiol. 52, 925–931 (2004)
Stockum, A., Lloyd, R. G. & Rudolph, C. J. On the viability of Escherichia coli cells lacking DNA topoisomerase I. BMC Microbiol. 12, 26 (2012)
Stupina, V. A. & Wang, J. C. Viability of Escherichia coli topA mutants lacking DNA topoisomerase I. J. Biol. Chem. 280, 355–360 (2005)
Yao, N. Y., Georgescu, R. E., Finkelstein, J. & O’Donnell, M. E. Single-molecule analysis reveals that the lagging strand increases replisome processivity but slows replication fork progression. Proc. Natl Acad. Sci. USA 106, 13236–13241 (2009)
Tanner, N. A. et al. Real-time single-molecule observation of rolling-circle DNA replication. Nucleic Acids Res. 37, e27 (2009)
O’Donnell, M. E. & Kornberg, A. Complete replication of templates by Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem. 260, 12884–12889 (1985)
McHenry, C. S. DNA replicases from a bacterial perspective. Annu. Rev. Biochem. 80, 403–436 (2011)
Wu, C. A., Zechner, E. L., Reems, J. A., McHenry, C. S. & Marians, K. J. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. V. Primase action regulates the cycle of Okazaki fragment synthesis. J. Biol. Chem. 267, 4074–4083 (1992)
Marko, J. F. & Siggia, E. D. Stretching DNA. Macromolecules 28, 8759–8770 (1995)
Dorman, C. J. DNA supercoiling and bacterial gene expression. Sci. Prog. 89, 151–166 (2006)
Travers, A. & Muskhelishvili, G. A common topology for bacterial and eukaryotic transcription initiation? EMBO Rep. 8, 147–151 (2007)
Benyajati, C. & Worcel, A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell 9, 393–407 (1976)
McInerney, P. & O’Donnell, M. Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J. Biol. Chem. 279, 21543–21551 (2004)
Yao, N., Hurwitz, J. & O’Donnell, M. Dynamics of beta and proliferating cell nuclear antigen sliding clamps in traversing DNA secondary structure. J. Biol. Chem. 275, 1421–1432 (2000)
Onrust, R., Finkelstein, J., Turner, J., Naktinis, V. & O’Donnell, M. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. III. Interface between two polymerases and the clamp loader. J. Biol. Chem. 270, 13366–13377 (1995)
Bailey, S., Wing, R. A. & Steitz, T. A. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell 126, 893–904 (2006)
Georgescu, R. E. et al. Mechanism of polymerase collision release from sliding clamps on the lagging strand. EMBO J. 28, 2981–2991 (2009)
Acknowledgements
We thank L. Langston, A. Libchaber and B. Michel for suggestions on the manuscript. We are also grateful to the National Institutes of Health (GM 38839) for supporting this work.
Author information
Authors and Affiliations
Contributions
I.K. and M.O.D. conceived the project, I.K. and R.G. performed experiments, and I.K., R.G. and M.O.D. designed the experiments, analysed data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-9. (PDF 2349 kb)
Rights and permissions
About this article
Cite this article
Kurth, I., Georgescu, R. & O'Donnell, M. A solution to release twisted DNA during chromosome replication by coupled DNA polymerases. Nature 496, 119–122 (2013). https://doi.org/10.1038/nature11988
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11988
This article is cited by
-
Effects of DNA supercoiling on chromatin architecture
Biophysical Reviews (2016)
-
Effects of DNA supercoiling on chromatin architecture
Biophysical Reviews (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.