Abstract
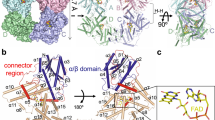
The cryptochrome (CRY) flavoproteins act as blue-light receptors in plants and insects, but perform light-independent functions at the core of the mammalian circadian clock. To drive clock oscillations, mammalian CRYs associate with the Period proteins (PERs) and together inhibit the transcription of their own genes. The SCFFBXL3 ubiquitin ligase complex controls this negative feedback loop by promoting CRY ubiquitination and degradation. However, the molecular mechanisms of their interactions and the functional role of flavin adenine dinucleotide (FAD) binding in CRYs remain poorly understood. Here we report crystal structures of mammalian CRY2 in its apo, FAD-bound and FBXL3–SKP1-complexed forms. Distinct from other cryptochromes of known structures, mammalian CRY2 binds FAD dynamically with an open cofactor pocket. Notably, the F-box protein FBXL3 captures CRY2 by simultaneously occupying its FAD-binding pocket with a conserved carboxy-terminal tail and burying its PER-binding interface. This novel F-box-protein–substrate bipartite interaction is susceptible to disruption by both FAD and PERs, suggesting a new avenue for pharmacological targeting of the complex and a multifaceted regulatory mechanism of CRY ubiquitination.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Chaves, I. et al. The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 62, 335–364 (2011)
Oztürk, N. et al. Structure and function of animal cryptochromes. Cold Spring Harb. Symp. Quant. Biol. 72, 119–131 (2007)
Lin, C. & Shalitin, D. Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54, 469–496 (2003)
Yuan, Q., Metterville, D., Briscoe, A. D. & Reppert, S. M. Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol. Biol. Evol. 24, 948–955 (2007)
Zhu, H. et al. The two CRYs of the butterfly. Curr. Biol. 15, R953–R954 (2005); erratum. 16, 730 (2006)
Stanewsky, R. et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692 (1998)
van der Horst, G. T. et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 (1999)
Griffin, E. A., Staknis, D. & Weitz, C. J. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286, 768–771 (1999)
Reppert, S. M. & Weaver, D. R. Coordination of circadian timing in mammals. Nature 418, 935–941 (2002)
Shearman, L. P. et al. Interacting molecular loops in the mammalian circadian clock. Science 288, 1013–1019 (2000)
Lee, C., Etchegaray, J. P., Cagampang, F. R., Loudon, A. S. & Reppert, S. M. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855–867 (2001)
Dibner, C., Schibler, U. & Albrecht, U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72, 517–549 (2010)
Green, C. B., Takahashi, J. S. & Bass, J. The meter of metabolism. Cell 134, 728–742 (2008)
Bass, J. & Takahashi, J. S. Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010)
Asher, G. & Schibler, U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 13, 125–137 (2011)
Yang, H. Q., Tang, R. H. & Cashmore, A. R. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13, 2573–2587 (2001)
Wang, H., Ma, L. G., Li, J. M., Zhao, H. Y. & Deng, X. W. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294, 154–158 (2001)
Peschel, N., Chen, K. F., Szabo, G. & Stanewsky, R. Light-dependent interactions between the Drosophila circadian clock factors Cryptochrome, Jetlag, and Timeless. Curr. Biol. 19, 241–247 (2009)
Koh, K., Zheng, X. & Sehgal, A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science 312, 1809–1812 (2006)
Zoltowski, B. D. et al. Structure of full-length Drosophila cryptochrome. Nature 480, 396–399 (2011)
Liu, B., Liu, H., Zhong, D. & Lin, C. Searching for a photocycle of the cryptochrome photoreceptors. Curr. Opin. Plant Biol. 13, 578–586 (2010)
Busino, L. et al. SCFFBXL3controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316, 900–904 (2007)
Godinho, S. I. et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316, 897–900 (2007)
Siepka, S. M. et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of Cryptochrome and Period gene expression. Cell 129, 1011–1023 (2007)
Lamia, K. A. et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326, 437–440 (2009)
Hirota, T. et al. Identification of small molecule activators of cryptochrome. Science 337, 1094–1097 (2012)
Müller, M. & Carell, T. Structural biology of DNA photolyases and cryptochromes. Curr. Opin. Struct. Biol. 19, 277–285 (2009)
Hitomi, K. et al. Functional motifs in the (6–4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes. Proc. Natl Acad. Sci. USA 106, 6962–6967 (2009)
Brautigam, C. A. et al. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 101, 12142–12147 (2004)
Maul, M. J. et al. Crystal structure and mechanism of a DNA (6–4) photolyase. Angew. Chem. Int. Edn Engl. 47, 10076–10080 (2008)
Park, H. W., Kim, S. T., Sancar, A. & Deisenhofer, J. Crystal structure of DNA photolyase from Escherichia coli. Science 268, 1866–1872 (1995)
Ozber, N. et al. Identification of two amino acids in the C-terminal domain of mouse CRY2 essential for PER2 interaction. BMC Mol. Biol. 11, 69 (2010)
Chaves, I. et al. Functional evolution of the photolyase/cryptochrome protein family: importance of the C terminus of mammalian CRY1 for circadian core oscillator performance. Mol. Cell. Biol. 26, 1743–1753 (2006)
Yagita, K. et al. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 21, 1301–1314 (2002)
Chen, R. et al. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol. Cell 36, 417–430 (2009)
Sanada, K., Harada, Y., Sakai, M., Todo, T. & Fukada, Y. Serine phosphorylation of mCRY1 and mCRY2 by mitogen-activated protein kinase. Genes Cells 9, 697–708 (2004)
Hao, B. et al. Structural basis of the Cks1-dependent recognition of p27Kip1 by the SCFSkp2 ubiquitin ligase. Mol. Cell 20, 9–19 (2005)
Tan, X. et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 (2007)
Otwinowski, Z. & Minor, W. In Methods in Enzymology Vol. 276 (eds Carter, C. W. & Sweet, R. M. ) 307–326 (Academic Press, 1997)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Collaborative Computational Project, number 4. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Acknowledgements
We thank the beamline staff of the Advanced Light Source at the University of California at Berkeley for help with data collection, and members of the Zheng laboratory for discussion. This work is supported by the Howard Hughes Medical Institute (N. Z. and M. P.), the National Institutes of Health (R01-CA107134 to N.Z., 5T32-HL007151 to L.B., and R01-GM057587, R37-CA-076584 and R21-CA161108 to M.P.), and the University of Washington (S.T.M. and M.F.B.).
Author information
Authors and Affiliations
Contributions
The protein purification and crystallization experiments were conceived by W.X., L.B., M.P. and N.Z., initiated by N.H.S., and conducted by W.X. W.X. and N.Z. determined and analysed the structures. FAD fluorescence and in vitro competition experiments were conceived by W.X., T.R.H. and N.Z., and conducted by W.X. and T.R.H. Mutational and binding studies, and stability analyses were conceived by L.B., M.P., W.X. and N.Z., and conducted by L.B. S.T.M. and M.F.B. conducted native mass spectrometry experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-12, Supplementary Methods, Supplementary Tables 1-2, a Supplementary Discussion and additional references. (PDF 7332 kb)
Rights and permissions
About this article
Cite this article
Xing, W., Busino, L., Hinds, T. et al. SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496, 64–68 (2013). https://doi.org/10.1038/nature11964
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11964
This article is cited by
-
Cryptochrome 2 acetylation attenuates its antiproliferative effect in breast cancer
Cell Death & Disease (2023)
-
Decoupling PER phosphorylation, stability and rhythmic expression from circadian clock function by abolishing PER-CK1 interaction
Nature Communications (2022)
-
CRY2 interacts with CIS1 to regulate thermosensory flowering via FLM alternative splicing
Nature Communications (2022)
-
Discovery of a small molecule that selectively destabilizes Cryptochrome 1 and enhances life span in p53 knockout mice
Nature Communications (2022)
-
Melatonergic Receptors (Mt1/Mt2) as a Potential Additional Target of Novel Drugs for Depression
Neurochemical Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.