Abstract
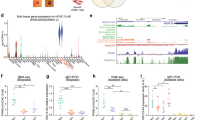
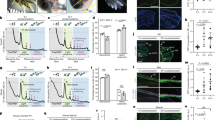
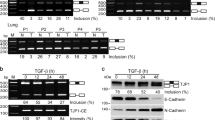
Post-transcriptional switches are flexible effectors of dynamic changes in gene expression1. Here we report a new post-transcriptional switch that dictates the spatiotemporal and mutually exclusive expression of two alternative gene products from a single transcript. Expression of primate-specific exonic microRNA-198 (miR-198)2, located in the 3′-untranslated region of follistatin-like 1 (FSTL1)3 messenger RNA, switches to expression of the linked open reading frame of FSTL1 upon wounding in a human ex vivo organ culture system. We show that binding of a KH-type splicing regulatory protein (KSRP, also known as KHSRP) to the primary transcript determines the fate of the transcript and is essential for the processing of miR-198: transforming growth factor-β signalling switches off miR-198 expression by downregulating KSRP, and promotes FSTL1 protein expression. We also show that FSTL1 expression promotes keratinocyte migration, whereas miR-198 expression has the opposite effect by targeting and inhibiting DIAPH1, PLAU and LAMC2. A clear inverse correlation between the expression pattern of FSTL1 (pro-migratory) and miR-198 (anti-migratory) highlights the importance of this regulatory switch in controlling context-specific gene expression to orchestrate wound re-epithelialization. The deleterious effect of failure of this switch is apparent in non-healing chronic diabetic ulcers, in which expression of miR-198 persists, FSTL1 is absent, and keratinocyte migration, re-epithelialization and wound healing all fail to occur.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Anderson, P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nature Rev. Immunol. 10, 24–35 (2010)
Hinske, L. C., Galante, P. A., Kuo, W. P. & Ohno-Machado, L. A potential role for intragenic miRNAs on their hosts’ interactome. BMC Genomics 11, 533 (2010)
Shibanuma, M., Mashimo, J., Mita, A., Kuroki, T. & Nose, K. Cloning from a mouse osteoblastic cell line of a set of transforming-growth-factor-β1-regulated genes, one of which seems to encode a follistatin-related polypeptide. Eur. J. Biochem. 217, 13–19 (1993)
Gurtner, G. C., Werner, S., Barrandon, Y. & Longaker, M. T. Wound repair and regeneration. Nature 453, 314–321 (2008)
Blakytny, R. & Jude, E. The molecular biology of chronic wounds and delayed healing in diabetes. Diabet. Med. 23, 594–608 (2006)
Banerjee, J., Chan, Y. C. & Sen, C. K. MicroRNAs in skin and wound healing. Physiol. Genomics 43, 543–556 (2011)
Tan, S., Li, R., Ding, K., Lobie, P. E. & Zhu, T. miR-198 inhibits migration and invasion of hepatocellular carcinoma cells by targeting the HGF/c-MET pathway. FEBS Lett. 585, 2229–2234 (2011)
Cai, X., Hagedorn, C. H. & Cullen, B. R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10, 1957–1966 (2004)
Hambrock, H. O. et al. Structural characterization of TSC-36/Flik: analysis of two charge isoforms. J. Biol. Chem. 279, 11727–11735 (2004)
Kroeze, K. L. et al. Autocrine regulation of re-epithelialization after wounding by chemokine receptors CCR1, CCR10, CXCR1, CXCR2, and CXCR3. J. Invest. Dermatol. 132, 216–225 (2012)
He, Y., Esser, P., Heinemann, A., Bruckner-Tuderman, L. & Has, C. Kindlin-1 and -2 have overlapping functions in epithelial cells implications for phenotype modification. Am. J. Pathol. 178, 975–982 (2011)
Freedberg, I. M., Tomic-Canic, M., Komine, M. & Blumenberg, M. Keratins and the keratinocyte activation cycle. J. Invest. Dermatol. 116, 633–640 (2001)
Lund, L. R. et al. Plasminogen activation independent of uPA and tPA maintains wound healing in gene-deficient mice. EMBO J. 25, 2686–2697 (2006)
Brandt, D. T. et al. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J. Cell Biol. 178, 193–200 (2007)
Frank, D. E. & Carter, W. G. Laminin 5 deposition regulates keratinocyte polarization and persistent migration. J. Cell Sci. 117, 1351–1363 (2004)
Trabucchi, M. et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459, 1010–1014 (2009)
Wang, B. et al. TGFβ-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene 29, 1787–1797 (2010)
Jude, E. B., Blakytny, R., Bulmer, J., Boulton, A. J. & Ferguson, M. W. Transforming growth factor-beta 1, 2, 3 and receptor type I and II in diabetic foot ulcers. Diabet. Med. 19, 440–447 (2002)
Pastar, I. et al. Attenuation of the transforming growth factor β-signaling pathway in chronic venous ulcers. Mol. Med. 16, 92–101 (2010)
Usui, M. L. et al. Morphological evidence for the role of suprabasal keratinocytes in wound reepithelialization. Wound Repair Regen. 13, 468–479 (2005)
Ridley, A. J. et al. Cell migration: integrating signals from front to back. Science 302, 1704–1709 (2003)
Daniel, R. J. & Groves, R. W. Increased migration of murine keratinocytes under hypoxia is mediated by induction of urokinase plasminogen activator. J. Invest. Dermatol. 119, 1304–1309 (2002)
Marutsuka, K., Woodcock-Mitchell, J., Sakamoto, T., Sobel, B. E. & Fujii, S. Pathogenetic implications of hyaluronan-induced modification of vascular smooth muscle cell fibrinolysis in diabetes. Coron. Artery Dis. 9, 177–184 (1998)
Zorio, E. et al. Fibrinolysis: the key to new pathogenetic mechanisms. Curr. Med. Chem. 15, 923–929 (2008)
Usui, M. L., Mansbridge, J. N., Carter, W. G., Fujita, M. & Olerud, J. E. Keratinocyte migration, proliferation, and differentiation in chronic ulcers from patients with diabetes and normal wounds. J. Histochem. Cytochem. 56, 687–696 (2008)
Stojadinovic, O. et al. Molecular pathogenesis of chronic wounds: the role of beta-catenin and c-myc in the inhibition of epithelialization and wound healing. Am. J. Pathol. 167, 59–69 (2005)
Jeffcoate, W. J. & Harding, K. G. Diabetic foot ulcers. Lancet 361, 1545–1551 (2003)
Brem, H. & Tomic-Canic, M. Cellular and molecular basis of wound healing in diabetes. J. Clin. Invest. 117, 1219–1222 (2007)
Acknowledgements
This work was supported by an A*STAR Investigatorship award to P.S., the Biomedical Research Council of Singapore and the Skin Biology Cluster Platform, A*STAR. We thank T. Kamala, S. Nama, M. Hisyam and C. Vaz for experimental support and B. Knowles for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
G.M.S. and J.E.A.C. performed most of the experiments; F.E.G. and D.P.L. helped with immunohistochemistry; K.L., T.C.L. and S.S. assisted in procurement of patient samples; V.T. helped in microarray data analysis; E.B.L. assisted in experimental design and contributed to writing the manuscript; and P.S. designed experiments, supervised this work and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-17 and lists of primers a-c. (PDF 1176 kb)
Live cell imaging of scratch wound assay in keratinocytes over-expressing miR-198
Keratinocytes over-expressing miR-198 were plated as a monolayer and subjected to scratch wound assay. Video represents time lapse imaging performed every one hour after wounding for a period of 48 hours. (MOV 4840 kb)
Live cell imaging of scratch wound assay in keratinocytes over-expressing control miRNA
Keratinocytes over-expressing control miRNA were plated as a monolayer and subjected to scratch wound assay. Video represents time lapse imaging performed every one hour after wounding for a period of 48 hours. (MOV 4832 kb)
Rights and permissions
About this article
Cite this article
Sundaram, G., Common, J., Gopal, F. et al. ‘See-saw’ expression of microRNA-198 and FSTL1 from a single transcript in wound healing. Nature 495, 103–106 (2013). https://doi.org/10.1038/nature11890
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11890
This article is cited by
-
Retro-miRs: novel and functional miRNAs originating from mRNA retrotransposition
Mobile DNA (2023)
-
Follistatin-like 1 and its paralogs in heart development and cardiovascular disease
Heart Failure Reviews (2022)
-
Follistatin-Like 1 Induces the Activation of Type 2 Innate Lymphoid Cells to Promote Airway Inflammation in Asthma
Inflammation (2022)
-
MicroRNA-200b/c-3p regulate epithelial plasticity and inhibit cutaneous wound healing by modulating TGF-β-mediated RAC1 signaling
Cell Death & Disease (2020)
-
Fstl1/DIP2A/MGMT signaling pathway plays important roles in temozolomide resistance in glioblastoma
Oncogene (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.