Abstract
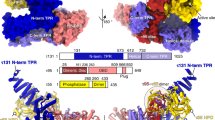
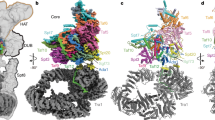
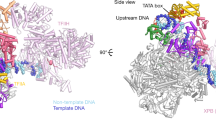
The initiation of gene transcription by RNA polymerase II is regulated by a plethora of proteins in human cells. The first general transcription factor to bind gene promoters is transcription factor IID (TFIID). TFIID triggers pre-initiation complex formation, functions as a coactivator by interacting with transcriptional activators and reads epigenetic marks1,2,3. TFIID is a megadalton-sized multiprotein complex composed of TATA-box-binding protein (TBP) and 13 TBP-associated factors (TAFs)3. Despite its crucial role, the detailed architecture and assembly mechanism of TFIID remain elusive. Histone fold domains are prevalent in TAFs, and histone-like tetramer and octamer structures have been proposed in TFIID4,5,6. A functional core-TFIID subcomplex was revealed in Drosophila nuclei, consisting of a subset of TAFs (TAF4, TAF5, TAF6, TAF9 and TAF12)7. These core subunits are thought to be present in two copies in holo-TFIID, in contrast to TBP and other TAFs that are present in a single copy8, conveying a transition from symmetry to asymmetry in the TFIID assembly pathway. Here we present the structure of human core-TFIID determined by cryo-electron microscopy at 11.6 Å resolution. Our structure reveals a two-fold symmetric, interlaced architecture, with pronounced protrusions, that accommodates all conserved structural features of the TAFs including the histone folds. We further demonstrate that binding of one TAF8–TAF10 complex breaks the original symmetry of core-TFIID. We propose that the resulting asymmetric structure serves as a functional scaffold to nucleate holo-TFIID assembly, by accreting one copy each of the remaining TAFs and TBP.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Data deposits
The cryo-EM maps have been deposited in the 3D-EM database (EMBL-European Bioinformatics Institute). EMBD accession codes are EMD-2229 (3TAF), EMD-2230 (core-TFIID) and EMD-2231 (7TAF).
References
Papai, G. et al. TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature 465, 956–960 (2010)
Müller, F., Zaucker, A. & Tora, L. Developmental regulation of transcription initiation: more than just changing the actors. Curr. Opin. Genet. Dev. 20, 533–540 (2010)
Papai, G., Weil, P. A. & Schultz, P. New insights into the function of transcription factor TFIID from recent structural studies. Curr. Opin. Genet. Dev. 21, 219–224 (2011)
Xie, X. et al. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature 380, 316–322 (1996)
Hoffmann, A. et al. A histone octamer-like structure within TFIID. Nature 380, 356–359 (1996)
Selleck, W. et al. A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nature Struct. Biol. 8, 695–700 (2001)
Wright, K. J., Marr, M. T., II & Tjian, R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc. Natl Acad. Sci. USA 103, 12347–12352 (2006)
Sanders, S. L., Garbett, K. A. & Weil, P. A. Molecular characterization of Saccharomyces cerevisiae TFIID. Mol. Cell. Biol. 22, 6000–6013 (2002)
Andel, F., III, Ladurner, A. G., Inouye, C., Tjian, R. & Nogales, E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 286, 2153–2156 (1999)
Brand, M., Leurent, C., Mallouh, V., Tora, L. & Schultz, P. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 286, 2151–2153 (1999)
Grob, P. et al. Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure 14, 511–520 (2006)
Cler, E., Papai, G., Schultz, P. & Davidson, I. Recent advances in understanding the structure and function of general transcription factor TFIID. Cell. Mol. Life Sci. 66, 2123–2134 (2009)
Papai, G. et al. Mapping the initiator binding Taf2 subunit in the structure of hydrated yeast TFIID. Structure 17, 363–373 (2009)
Dubrovskaya, V. et al. Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIF beta (RAP30) and incorporation into the TFIID complex. EMBO J. 15, 3702–3712 (1996)
Bhattacharya, S., Takada, S. & Jacobson, R. H. Structural analysis and dimerization potential of the human TAF5 subunit of TFIID. Proc. Natl Acad. Sci. USA 104, 1189–1194 (2007)
Romier, C. et al. Crystal structure, biochemical and genetic characterization of yeast and E. cuniculi TAF(II)5 N-terminal domain: implications for TFIID assembly. J. Mol. Biol. 368, 1292–1306 (2007)
Scheer, E., Delbac, F., Tora, L., Moras, D. & Romier, C. TFIID TAF6–TAF9 complex formation involves the HEAT repeat-containing C-terminal domain of TAF6 and is modulated by TAF5. J. Biol. Chem. 287, 27580–27592 (2012)
Fitzgerald, D. J. et al. Multiprotein expression strategy for structural biology of eukaryotic complexes. Structure 15, 275–279 (2007)
Werten, S. et al. Crystal structure of a subcomplex of human transcription factor TFIID formed by TATA binding protein-associated factors hTAF4 (hTAFII135) and hTAF12 (hTAFII20). J. Biol. Chem. 277, 45502–45509 (2002)
Wang, X. et al. Conserved region I of human coactivator TAF4 binds to a short hydrophobic motif present in transcriptional regulators. Proc. Natl Acad. Sci. USA 104, 7839–7844 (2007)
Leurent, C. et al. Mapping histone fold TAFs within yeast TFIID. EMBO J. 21, 3424–3433 (2002)
Leurent, C. et al. Mapping key functional sites within yeast TFIID. EMBO J. 23, 719–727 (2004)
Soutoglou, E. et al. The nuclear import of TAF10 is regulated by one of its three histone fold domain-containing interaction partners. Mol. Cell. Biol. 25, 4092–4104 (2005)
Demény, M. A. et al. Identification of a small TAF complex and its role in the assembly of TAF-containing complexes. PLoS ONE 2, e316 (2007)
Burke, T. W. & Kadonaga, J. T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11, 3020–3031 (1997)
Timmers, H. T. M. & Tora, L. SAGA unveiled. Trends Biochem. Sci. 30, 7–10 (2005)
Mengus, G. et al. TAF4 inactivation in embryonic fibroblasts activates TGFβ signaling and autocrine growth. EMBO J. 24, 2753–2767 (2005)
Liu, W.-L. et al. Structural changes in TAF4b–TFIID correlate with promoter selectivity. Mol. Cell 29, 81–91 (2008)
Bell, B., Scheer, E. & Tora, L. Identification of hTAFII80δ links apoptotic signaling pathways to transcription factor TFIID function. Mol. Cell 8, 591–600 (2001)
Chen, Z. & Manley, J. L. In vivo functional analysis of the histone 3-like TAF9 and a TAF9-related factor, TAF9L. J. Biol. Chem. 278, 35172–35183 (2003)
Acknowledgements
We thank all members of the Berger, Schultz and Tora laboratories for advice and discussions. We are grateful to C. Romier for providing the X-ray structure of the TAF6 C-terminal domain before publication. We thank J. Demmers for mass spectrometric analyses and F. Grosveld for discussions. The EMBL, IBS and IGBMC core facilities are acknowledged for services. We are indebted to G. Schoehn for maintaining the electron microscopes in Grenoble. T. J. Richmond is acknowledged for advice and support. C.B. is a fellow of the joint European Commission (EC)/EMBL interdisciplinary research opportunities program (EIPOD). C.S. is recipient of a European Research Council (ERC) Starting Grant and an Agence Nationale de la Recherche (ANR) Jeunes Chercheuses award. I.B. acknowledges support from the EC Marie Curie Action and the EC Framework Programme (FP) 7 projects INSTRUCT, PCUBE, BioSTRUCT-X, 4D-CellFate and ComplexINC. P.S. acknowledges support from the Institut National de la Santé et de la Recherche Médicale (INSERM), the Centre National pour la Recherche Scientifique (CNRS), the Association pour la Recherche sur le Cancer (ARC) and the Fondation pour la Recherche Médicale (FRM). This work was supported by the ANR Projets Blancs puzzle-fit (to P.S.), ChromAct (to P.S. and L.T.) and TFIID-Complexes (to L.T., P.S. and I.B.).
Author information
Authors and Affiliations
Contributions
P.S., L.T. and I.B. designed the study; C.B., F.G. and I.B. implemented the MultiBac system; C.B., F.G. and M.C. produced, purified and characterized all TAF complexes; C.S. implemented gradient centrifugation and GraFix, analysed negative-stain electron microscopy data and calculated two-dimensional class averages; G.P. and P.S. carried out random conical tilt experiments, collected and analysed cryo-EM data of all complexes, and calculated and refined the electron microscopy densities; G.P. prepared the core-TFIID molecular structure by fitting crystal coordinates and homology models; E.S., L.T. and P.P. prepared and analysed endogenous TFIID for protein content. L.T. provided the anti-TAF10 antibody (mAb6TA). P.S., L.T. and I.B. supervised the work. G.P., C.B., P.S., L.T. and I.B. prepared the figures and wrote the manuscript together.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Tables 1-3, Supplementary Text, Supplementary Figures 1-15 and Supplementary References. (PDF 4507 kb)
Human core-TFIID structure in an animated view
This video shows the core-TFIID cryo-EM structure (grey mesh), with the assigned TAF domains, crystal structures and homology models superimposed (color code as in Figure 1). Core-TFIID rotates around 360° in the video, illustrating the structure (MPEG-4 (H.264)). (MP4 13944 kb)
Rights and permissions
About this article
Cite this article
Bieniossek, C., Papai, G., Schaffitzel, C. et al. The architecture of human general transcription factor TFIID core complex. Nature 493, 699–702 (2013). https://doi.org/10.1038/nature11791
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11791
This article is cited by
-
Hierarchical TAF1-dependent co-translational assembly of the basal transcription factor TFIID
Nature Structural & Molecular Biology (2023)
-
Loss of TAF8 causes TFIID dysfunction and p53-mediated apoptotic neuronal cell death
Cell Death & Differentiation (2022)
-
GoldenBac: a simple, highly efficient, and widely applicable system for construction of multi-gene expression vectors for use with the baculovirus expression vector system
BMC Biotechnology (2020)
-
TAF1, associated with intellectual disability in humans, is essential for embryogenesis and regulates neurodevelopmental processes in zebrafish
Scientific Reports (2019)
-
Organization and regulation of gene transcription
Nature (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.