Abstract
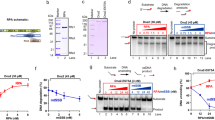
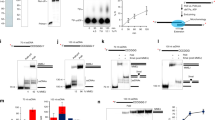
Replicative DNA helicases generally unwind DNA as a single hexamer that encircles and translocates along one strand of the duplex while excluding the complementary strand (known as steric exclusion). By contrast, large T antigen, the replicative DNA helicase of the simian virus 40 (SV40), is reported to function as a pair of stacked hexamers that pumps double-stranded DNA through its central channel while laterally extruding single-stranded DNA. Here we use single-molecule and ensemble assays to show that large T antigen assembled on the SV40 origin unwinds DNA efficiently as a single hexamer that translocates on single-stranded DNA in the 3′-to-5′ direction. Unexpectedly, large T antigen unwinds DNA past a DNA–protein crosslink on the translocation strand, suggesting that the large T antigen ring can open to bypass bulky adducts. Together, our data underscore the profound conservation among replicative helicase mechanisms, and reveal a new level of plasticity in the interactions of replicative helicases with DNA damage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Patel, S. S. & Picha, K. M. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69, 651–697 (2000)
Enemark, E. J. & Joshua-Tor, L. On helicases and other motor proteins. Curr. Opin. Struct. Biol. 18, 243–257 (2008)
Egelman, E. H., Yu, X., Wild, R., Hingorani, M. M. & Patel, S. S. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl Acad. Sci. USA 92, 3869–3873 (1995)
Kaplan, D. L. & O’Donnell, M. Twin DNA pumps of a hexameric helicase provide power to simultaneously melt two duplexes. Mol. Cell 15, 453–465 (2004)
Fu, Y. V. et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146, 931–941 (2011)
Kaplan, D. L., Davey, M. J. & O’Donnell, M. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J. Biol. Chem. 278, 49171–49182 (2003)
Fanning, E. & Zhao, K. SV40 DNA replication: from the A gene to a nanomachine. Virology 384, 352–359 (2009)
Fanning, E., Zhao, X. & Jiang, X. in DNA Tumor Viruses (eds Damania, B. & Pipas, J. M. ) 1–24 (Springer US, 2009)
Wessel, R., Schweizer, J. & Stahl, H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J. Virol. 66, 804–815 (1992)
Weisshart, K. et al. Two regions of simian virus 40 large T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J. Virol. 73, 2201–2211 (1999)
Barbaro, B. A., Sreekumar, K. R., Winters, D. R., Prack, A. E. & Bullock, P. A. Phosphorylation of simian virus 40 large T antigen on Thr 124 selectively promotes double-hexamer formation on subfragments of the viral core origin. J. Virol. 74, 8601–8613 (2000)
Smelkova, N. V. & Borowiec, J. A. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J. Virol. 71, 8766–8773 (1997)
Alexandrov, A. I., Botchan, M. R. & Cozzarelli, N. R. Characterization of simian virus 40 T-antigen double hexamers bound to a replication fork. J. Biol. Chem. 277, 44886–44897 (2002)
SenGupta, D. J. & Borowiec, J. A. Strand-specific recognition of a synthetic DNA replication fork by the SV40 large tumor antigen. Science 256, 1656–1661 (1992)
Morris, P. D. et al. Hepatitis C virus NS3 and simian virus 40 large T antigen helicases displace streptavidin from 5′-biotinylated oligonucleotides but not from 3′-biotinylated oligonucleotides: evidence for directional bias in translocation on single-stranded DNA. Biochemistry 41, 2372–2378 (2002)
Goetz, G. S., Dean, F. B., Hurwitz, J. & Matson, S. W. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J. Biol. Chem. 263, 383–392 (1988)
Enemark, E. J. & Joshua-Tor, L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442, 270–275 (2006)
Li, D. et al. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423, 512–518 (2003)
Gai, D., Zhao, R., Li, D., Finkielstein, C. V. & Chen, X. S. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119, 47–60 (2004)
Gomez-Lorenzo, M. G. et al. Large large T antigen on the simian virus 40 origin of replication: a 3D snapshot prior to DNA replication. EMBO J. 22, 6205–6213 (2003)
Cuesta, I. et al. Conformational rearrangements of SV40 large large T antigen during early replication events. J. Mol. Biol. 397, 1276–1286 (2010)
Borowiec, J. A. & Hurwitz, J. ATP stimulates the binding of simian virus 40 (SV40) large tumor antigen to the SV40 origin of replication. Proc. Natl Acad. Sci. USA 85, 64–68 (1988)
Sclafani, R. A., Fletcher, R. J. & Chen, X. S. Two heads are better than one: regulation of DNA replication by hexameric helicases. Genes Dev. 18, 2039–2045 (2004)
Takahashi, T. S., Wigley, D. B. & Walter, J. C. Pumps, paradoxes and ploughshares: mechanism of the MCM2–7 DNA helicase. Trends Biochem. Sci. 30, 437–444 (2005)
Yardimci, H., Loveland, A. B., Habuchi, S., van Oijen, A. M. & Walter, J. C. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol. Cell 40, 834–840 (2010)
Yardimci, H., Loveland, A. B., van Oijen, A. M. & Walter, J. C. Single-molecule analysis of DNA replication in Xenopus egg extracts. Methods 57, 179–186 (2012)
Stillman, B. W. & Gluzman, Y. Replication and supercoiling of simian virus 40 DNA in cell extracts from human cells. Mol. Cell. Biol. 5, 2051–2060 (1985)
Wobbe, C. R., Dean, F., Weissbach, L. & Hurwitz, J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc. Natl Acad. Sci. USA 82, 5710–5714 (1985)
Bullock, P. A., Seo, Y. S. & Hurwitz, J. Initiation of simian virus 40 DNA synthesis in vitro. Mol. Cell. Biol. 11, 2350–2361 (1991)
Murakami, Y. & Hurwitz, J. Functional interactions between SV40 large T antigen and other replication proteins at the replication fork. J. Biol. Chem. 268, 11008–11017 (1993)
Kim, S., Dallmann, H. G., McHenry, C. S. & Marians, K. J. Coupling of a replicative polymerase and helicase: a τ-DnaB interaction mediates rapid replication fork movement. Cell 84, 643–650 (1996)
Stano, N. M. et al. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature 435, 370–373 (2005)
Chen, L. et al. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry 30, 11018–11025 (1991)
Seinsoth, S., Uhlmann-Schiffler, H. & Stahl, H. Bidirectional DNA unwinding by a ternary complex of large T antigen, nucleolin and topoisomerase I. EMBO Rep. 4, 263–268 (2003)
Barker, S., Weinfeld, M. & Murray, D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 589, 111–135 (2005)
Anand, R. P. et al. Overcoming natural replication barriers: differential helicase requirements. Nucleic Acids Res. 40, 1091–1105 (2011)
Wu, C., Roy, R. & Simmons, D. T. Role of single-stranded DNA binding activity of large T antigen in simian virus 40 DNA replication. J. Virol. 75, 2839–2847 (2001)
Eki, T., Matsumoto, T., Murakami, Y. & Hurwitz, J. The replication of DNA containing the simian virus 40 origin by the monopolymerase and dipolymerase systems. J. Biol. Chem. 267, 7284–7294 (1992)
Ishimi, Y., Claude, A., Bullock, P. & Hurwitz, J. Complete enzymatic synthesis of DNA containing the SV40 origin of replication. J. Biol. Chem. 263, 19723–19733 (1988)
Wold, M. S., Li, J. J. & Kelly, T. J. Initiation of simian virus 40 DNA replication in vitro: large-tumor-antigen- and origin-dependent unwinding of the template. Proc. Natl Acad. Sci. USA 84, 3643–3647 (1987)
Wang, I.-N. Lysis timing and bacteriophage fitness. Genetics 172, 17–26 (2005)
Thomason, L. C., Oppenheim, A. B. & Court, D. L. Modifying bacteriophage lambda with recombineering. Methods Mol Biol. 501, 239–251 (2009)
Kuhn, H. & Frank-Kamenetskii, M. D. Labeling of unique sequences in double-stranded DNA at sites of vicinal nicks generated by nicking endonucleases. Nucleic Acids Res. 36, e40 (2008)
Loparo, J. J., Kulczyk, A. W., Richardson, C. C. & van Oijen, A. M. Simultaneous single-molecule measurements of phage T7 replisome composition and function reveal the mechanism of polymerase exchange. Proc. Natl Acad. Sci. USA 108, 3584–3589 (2011)
MacMillan, A. M., Chen, L. & Verdine, G. L. Synthesis of an oligonucleotide suicide substrate for DNA methyltransferases. J. Org. Chem. 57, 2989–2991 (1992)
Tanner, N. A. & van Oijen, A. M. in Single Molecule Tools, Part B:Super-Resolution, Particle Tracking, Multiparameter, and Force Based Methods (ed. Walter, N. G. ) 259–278 (Academic, 2010)
Acknowledgements
We thank C. Richardson for critical reading of the manuscript, and C. Etson for help in the preparation of λori DNA. J.C.W. was supported by grants from the National Institutes of Health (NIH) (GM62267 and HL098316) and American Cancer Society (ACS) (RSG0823401GMC). A.M.v.O. was supported by grants from the NIH (GM077248), ACS (RSG0823401GMC), and the Netherlands Organization for Scientific Research (NWO; Vici 680-47-607). J.H. was funded by NIH grant GM5 R01 GM034559. X.W. was a long-term postdoctoral fellow of the Human Frontier Science Program; D.Z.R. was supported by NIH grant GM086466.
Author information
Authors and Affiliations
Contributions
H.Y., A.M.v.O. and J.C.W. designed the experiments. H.Y. performed the experiments. X.W. and D.Z.R. supervised and assisted H.Y. for construction of λori DNA. A.B.L. made fluorescently tagged RPA. I.T. and J.H. provided RPA and large T antigen. H.Y., A.M.v.O. and J.C.W. interpreted the data and wrote the paper. X.W. and A.B.L. contributed equally.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-7. (PDF 591 kb)
Rights and permissions
About this article
Cite this article
Yardimci, H., Wang, X., Loveland, A. et al. Bypass of a protein barrier by a replicative DNA helicase. Nature 492, 205–209 (2012). https://doi.org/10.1038/nature11730
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11730
This article is cited by
-
Control of DNA replication in vitro using a reversible replication barrier
Nature Protocols (2024)
-
Three-dimensional super-resolution fluorescence imaging of DNA
Scientific Reports (2020)
-
Duplex DNA engagement and RPA oppositely regulate the DNA-unwinding rate of CMG helicase
Nature Communications (2020)
-
DNA translocation mechanism of the MCM complex and implications for replication initiation
Nature Communications (2019)
-
The mechanism of DNA unwinding by the eukaryotic replicative helicase
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.