Abstract
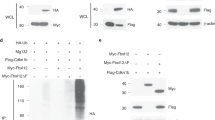
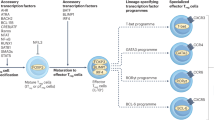
The differentiation of several T- and B-cell effector programs in the immune system is directed by signature transcription factors that induce rapid epigenetic remodelling. Here we report that promyelocytic leukaemia zinc finger (PLZF), the BTB-zinc finger (BTB-ZF) transcription factor directing the innate-like effector program of natural killer T-cell thymocytes1,2, is prominently associated with cullin 3 (CUL3), an E3 ubiquitin ligase previously shown to use BTB domain-containing proteins as adaptors for substrate binding3,4,5,6,7. PLZF transports CUL3 to the nucleus, where the two proteins are associated within a chromatin-modifying complex. Furthermore, PLZF expression results in selective ubiquitination changes of several components of this complex. CUL3 was also found associated with the BTB-ZF transcription factor BCL6, which directs the germinal-centre B cell and follicular T-helper cell programs. Conditional CUL3 deletion in mice demonstrated an essential role for CUL3 in the development of PLZF- and BCL6-dependent lineages. We conclude that distinct lineage-specific BTB-ZF transcription factors recruit CUL3 to alter the ubiquitination pattern of their associated chromatin-modifying complex. We propose that this new function is essential to direct the differentiation of several T- and B-cell effector programs, and may also be involved in the oncogenic role of PLZF and BCL6 in leukaemias and lymphomas8,9.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Savage, A. K. et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008)
Kovalovsky, D. et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nature Immunol. 9, 1055–1064 (2008)
Xu, L. et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425, 316–321 (2003)
Furukawa, M., He, Y. J., Borchers, C. & Xiong, Y. Targeting of protein ubiquitination by BTB–Cullin 3–Roc1 ubiquitin ligases. Nature Cell Biol. 5, 1001–1007 (2003)
Geyer, R., Wee, S., Anderson, S., Yates, J. & Wolf, D. A. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12, 783–790 (2003)
Pintard, L., Willems, A. & Peter, M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 23, 1681–1687 (2004)
Zimmerman, E. S., Schulman, B. A. & Zheng, N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 20, 714–721 (2010)
Basso, K. & Dalla-Favera, R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv. Immunol. 105, 193–210 (2010)
McConnell, M. J. & Licht, J. D. The PLZF gene of t(11;17)-associated APL. Curr. Top. Microbiol. Immunol. 313, 31–48 (2007)
Guidez, F. et al. RARα-PLZF overcomes PLZF-mediated repression of CRABPI, contributing to retinoid resistance in t(11;17) acute promyelocytic leukemia. Proc. Natl Acad. Sci. USA 104, 18694–18699 (2007)
Yasui, D., Miyano, M., Cai, S., Varga-Weisz, P. & Kohwi-Shigematsu, T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419, 641–645 (2002)
Cai, S., Lee, C. C. & Kohwi-Shigematsu, T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nature Genet. 38, 1278–1288 (2006)
Reddy, K. L., Zullo, J. M., Bertolino, E. & Singh, H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452, 243–247 (2008)
Zullo, J. M. et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 149, 1474–1487 (2012)
Ohnuki, H. et al. BAZF, a novel component of cullin3-based E3 ligase complex, mediates VEGFR and Notch cross-signaling in angiogenesis. Blood 119, 2688–2698 (2012)
Savage, A. K., Constantinides, M. G. & Bendelac, A. Promyelocytic leukemia zinc finger turns on the effector T cell program without requirement for agonist TCR signaling. J. Immunol. 186, 5801–5806 (2011)
Kwon, J. E. et al. BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J. Biol. Chem. 281, 12664–12672 (2006)
Wimuttisuk, W. & Singer, J. D. The Cullin3 ubiquitin ligase functions as a Nedd8-bound heterodimer. Mol. Biol. Cell 18, 899–909 (2007)
Errington, W. J. et al. Adaptor protein self-assembly drives the control of a Cullin-RING ubiquitin ligase. Structure 20, 1141–1153 (2012)
Crotty, S. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29, 621–663 (2011)
Hyjek, E., Chadburn, A., Liu, Y. F., Cesarman, E. & Knowles, D. M. BCL-6 protein is expressed in precursor T-cell lymphoblastic lymphoma and in prenatal and postnatal thymus. Blood 97, 270–276 (2001)
Braun, S. et al. The Cul4-Ddb1Cdt2 ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell 144, 41–54 (2011)
Wang, H. et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 (2004)
Bosch-Presegué, L. et al. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol. Cell 42, 210–223 (2011)
Du, Z. et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signal. 3, ra80 (2010)
Hernández-Muñoz, I. et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl Acad. Sci. USA 102, 7635–7640 (2005)
Hargreaves, D. C. & Crabtree, G. R. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 21, 396–420 (2011)
Sowa, M. E., Bennett, E. J., Gygi, S. P. & Harper, J. W. Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009)
Beisel, C. & Paro, R. Silencing chromatin: comparing modes and mechanisms. Nature Rev. Genet. 12, 123–135 (2011)
McEvoy, J. D., Kossatz, U., Malek, N. & Singer, J. D. Constitutive turnover of cyclin E by Cul3 maintains quiescence. Mol. Cell. Biol. 27, 3651–3666 (2007)
Jin, J., Ang, X. L., Shirogane, T. & Wade Harper, J. Identification of substrates for F-box proteins. Methods Enzymol. 399, 287–309 (2005)
Rush, J. et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nature Biotechnol. 23, 94–101 (2005)
Benlagha, K., Wei, D. G., Veiga, J., Teyton, L. & Bendelac, A. Characterization of the early stages in thymic NKT cell development. J. Exp. Med. 202, 485–492 (2005)
Acknowledgements
We thank A. Dinner, J. Licht, G. Prive, A. Ruthenburg, R. Sciammas, H. Singh and P. Wilson for discussions, J. C. Silva and M. Stokes for UbiScan analysis, C. Labno and V. Bindokas for help with confocal microscopy, and K. Block, L. Roach, D. Zabner, F. Meng and L. Bai for help with experiments. This work was supported by National Institute of Health (NIH) grants 5RO1GM082940 (J.D.S.) and RO1AI038339 (A.B.), and an Irvington Institute postdoctoral fellowship from the Cancer Research Institute (R.M.). A.B. is a Howard Hughes Medical Institute Investigator.
Author information
Authors and Affiliations
Contributions
R.M. designed the research, performed experiments and analysed data. M.P.S., S.T.S., A.M., M.G.C. and C.B.-V. performed experiments and analysed data. J.D.S. helped to design experiments and provided the Cul3fl/fl mice and CUL3 constructs. R.M. and A.B. co-wrote the paper. A.B. supervised the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-8, Supplementary Methods and a Supplementary Reference. (PDF 1858 kb)
Rights and permissions
About this article
Cite this article
Mathew, R., Seiler, M., Scanlon, S. et al. BTB-ZF factors recruit the E3 ligase cullin 3 to regulate lymphoid effector programs. Nature 491, 618–621 (2012). https://doi.org/10.1038/nature11548
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11548
This article is cited by
-
The dual roles of autophagy and the GPCRs-mediating autophagy signaling pathway after cerebral ischemic stroke
Molecular Brain (2022)
-
Leukemia/lymphoma-related factor (LRF) or osteoclast zinc finger protein (OCZF) overexpression promotes osteoclast survival by increasing Bcl-xl mRNA: A novel regulatory mechanism mediated by the RNA binding protein SAM68
Laboratory Investigation (2022)
-
The CRL3BTBD9 E3 ubiquitin ligase complex targets TNFAIP1 for degradation to suppress cancer cell migration
Signal Transduction and Targeted Therapy (2020)
-
The diverse roles of SPOP in prostate cancer and kidney cancer
Nature Reviews Urology (2020)
-
Cullin-3: Renal and Vascular Mechanisms Regulating Blood Pressure
Current Hypertension Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.