Abstract
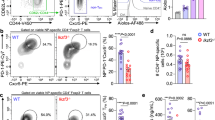
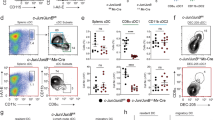
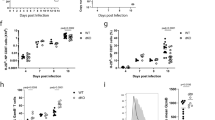
The AP1 transcription factor Batf3 is required for homeostatic development of CD8α+ classical dendritic cells that prime CD8 T-cell responses against intracellular pathogens. Here we identify an alternative, Batf3-independent pathway in mice for CD8α+ dendritic cell development operating during infection with intracellular pathogens and mediated by the cytokines interleukin (IL)-12 and interferon-γ. This alternative pathway results from molecular compensation for Batf3 provided by the related AP1 factors Batf, which also functions in T and B cells, and Batf2 induced by cytokines in response to infection. Reciprocally, physiological compensation between Batf and Batf3 also occurs in T cells for expression of IL-10 and CTLA4. Compensation among BATF factors is based on the shared capacity of their leucine zipper domains to interact with non-AP1 factors such as IRF4 and IRF8 to mediate cooperative gene activation. Conceivably, manipulating this alternative pathway of dendritic cell development could be of value in augmenting immune responses to vaccines.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Dorsey, M. J. et al. B-ATF: a novel human bZIP protein that associates with members of the AP-1 transcription factor family. Oncogene 11, 2255–2265 (1995)
Wagner, E. F. & Eferl, R. Fos/AP-1 proteins in bone and the immune system. Immunol. Rev. 208, 126–140 (2005)
Williams, K. L. et al. Characterization of murine BATF: a negative regulator of activator protein-1 activity in the thymus. Eur. J. Immunol. 31, 1620–1627 (2001)
Echlin, D. R., Tae, H. J., Mitin, N. & Taparowsky, E. J. B-ATF functions as a negative regulator of AP-1 mediated transcription and blocks cellular transformation by Ras and Fos. Oncogene 19, 1752–1763 (2000)
Schraml, B. U. et al. The AP-1 transcription factor Batf controls TH17 differentiation. Nature 460, 405–409 (2009)
Ise, W. et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nature Immunol. 12, 536–543 (2011)
Hildner, K. et al. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008)
Edelson, B. T. et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J. Exp. Med. 207, 823–836 (2010)
Betz, B. C. et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med. 207, 933–942 (2010)
Mashayekhi, M. et al. CD8a+ dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity 35, 249–259 (2011)
Bar-On, L. et al. CX3CR1+ CD8α+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc. Natl Acad. Sci. USA 107, 14745–14750 (2010)
Behnke, M. S. et al. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc. Natl Acad. Sci. USA 108, 9631–9636 (2011)
Ginhoux, F. et al. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206, 3115–3130 (2009)
Modlin, R. L. & Barnes, P. F. IL12 and the human immune response to mycobacteria. Res. Immunol. 146, 526–531 (1995)
den Haan, J. M., Lehar, S. M. & Bevan, M. J. CD8+ but not CD8− dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 192, 1685–1696 (2000)
Wilson, N. S. et al. Systemic activation of dendritic cells by Toll-like receptor ligands or malaria infection impairs cross-presentation and antiviral immunity. Nature Immunol. 7, 165–172 (2006)
Aliberti, J. et al. Essential role for ICSBP in the in vivo development of murine CD8α+ dendritic cells. Blood 101, 305–310 (2003)
Tailor, P., Tamura, T., Morse, H. C. & Ozato, K. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood 111, 1942–1945 (2008)
Edelson, B. T. et al. Batf3-dependent CD11blow/− peripheral dendritic cells are GM-CSF-independent and are not required for Th cell priming after subcutaneous immunization. PLoS ONE 6, e25660 (2011)
Marquis, J. F., Lacourse, R., Ryan, L., North, R. J. & Gros, P. Disseminated and rapidly fatal tuberculosis in mice bearing a defective allele at IFN regulatory factor 8. J. Immunol. 182, 3008–3015 (2009)
Jackson, J. T. et al. Id2 expression delineates differential checkpoints in the genetic program of CD8α+ and CD103+ dendritic cell lineages. EMBO J. 30, 2690–2704 (2011)
Edelson, B. T. et al. CD8a+ dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes. Immunity 35, 236–248 (2011)
Su, Z. Z. et al. Cloning and characterization of SARI (suppressor of AP-1, regulated by IFN). Proc. Natl Acad. Sci. USA 105, 20906–20911 (2008)
Ravasi, T. et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140, 744–752 (2010)
Brüstle, A. et al. The development of inflammatory TH17 cells requires interferon-regulatory factor 4. Nature Immunol. 8, 958–966 (2007)
Klein, U. et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nature Immunol. 7, 773–782 (2006)
Tamura, T. & Ozato, K. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J. Interferon Cytokine Res. 22, 145–152 (2002)
Sciammas, R. et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity 25, 225–236 (2006)
Glasmacher, E. et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science (in the press)
Li, P. et al. BATF–JUN is critical for IRF4-mediated transcription in T cells. Nature https://doi.org/10.1038/nature11530 (19 September 2012)
Brass, A. L., Zhu, A. Q. & Singh, H. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J. 18, 977–991 (1999)
Chen, L., Glover, J. N., Hogan, P. G., Rao, A. & Harrison, S. C. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature 392, 42–48 (1998)
Eisenbeis, C. F., Singh, H. & Storb, U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 9, 1377–1387 (1995)
Heng, T. S. & Painter, M. W. The Immunological Genome Project: networks of gene expression in immune cells. Nature Immunol. 9, 1091–1094 (2008)
Ranganath, S. et al. GATA-3-dependent enhancer activity in IL-4 gene regulation. J. Immunol. 161, 3822–3826 (1998)
Gorman, J. R. et al. The Igκ enhancer influences the ratio of Igκ versus Igλ B lymphocytes. Immunity 5, 241–252 (1996)
Iiizumi, S. et al. Simple one-week method to construct gene-targeting vectors: application to production of human knockout cell lines. Biotechniques 41, 311–316 (2006)
Edelson, B. T. et al. CD8a+ dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes. Immunity 35, 236–248 (2011)
Mashayekhi, M. et al. CD8a+ dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity 35, 249–259 (2011)
Edelson, B. T. et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J. Exp. Med. 207, 823–836 (2010)
Saeij, J. P., Boyle, J. P., Grigg, M. E., Arrizabalaga, G. & Boothroyd, J. C. Bioluminescence imaging of Toxoplasma gondii infection in living mice reveals dramatic differences between strains. Infect. Immun. 73, 695–702 (2005)
Hildner, K. et al. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008)
Matsushita, H. et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 482, 400–404 (2012)
Satpathy, A. T. et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 209, 1135–1152 (2012)
Schraml, B. U. et al. The AP-1 transcription factor Batf controls TH17 differentiation. Nature 460, 405–409 (2009)
Li, C. & Wong, W. H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA 98, 31–36 (2001)
Bending, D. et al. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J. Clin. Invest.. 119, 565–572 (2009)
Ise, W. et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nature Immunol. 12, 536–543 (2011)
Ranganath, S. et al. GATA-3-dependent enhancer activity in IL-4 gene regulation. J. Immunol. 161, 3822–3826 (1998)
Sedy, J. R. et al. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nature Immunol. 6, 90–98 (2005)
Kanbe, E. & Zhang, D. E. A simple and quick method to concentrate MSCV retrovirus. Blood Cells Mol. Dis. 33, 64–67 (2004)
Dignam, J. D., Lebovitz, R. M. & Roeder, R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 (1983)
Szabo, S. J., Gold, J. S., Murphy, T. L. & Murphy, K. M. Identification of cis-acting regulatory elements controlling interleukin-4 gene expression in T cells: roles for NF-Y and NF-ATc. Mol. Cell. Biol. 13, 4793–4805 (1993); erratum. 13, 5928 (1993)
Eisenbeis, C. F., Singh, H. & Storb, U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 9, 1377–1387 (1995)
Acknowledgements
This work was supported by the Howard Hughes Medical Institute, National Institutes of Health (AI076427-02) and Department of Defense (W81XWH-09-1-0185) (K.M.M.), the American Heart Association (12PRE8610005) (A.T.S.), German Research Foundation (AL 1038/1-1) (J.C.A), American Society of Hematology Scholar Award and Burroughs Welcome Fund Career Award for Medical Scientists (B.T.E.), and Cancer Research Institute predoctoral fellowship (W.-L.L.). We thank the ImmGen consortium34, M. White for blastocyst injections and generation of mouse chimaeras, the Alvin J. Siteman Cancer Center at Washington University School of Medicine for use of the Center for Biomedical Informatics and Multiplex Gene Analysis Genechip Core Facility. The Siteman Cancer Center is supported in part by the NCI Cancer Center Support Grant P30 CA91842. IL-12 was a gift from Pfizer.
Author information
Authors and Affiliations
Contributions
R.T., W.-L.L., T.L.M. and M.M. performed experiments with Batf−/−, Batf2−/−, Batf3−/− and double-knockout mice; T.L.M. made and analysed all BATF mutants; J.A.R. aided with EMSAs; W.KC, J.C.A. and A.T.S. were involved with microarray analysis and generation of mutant mice. J.C.A. was involved in generating of Batf2−/− mice. B.T.E. was involved with L. monocytogenes analysis; N.M.K was involved with cross-presentation analysis. X.W. was involved with bioinformatic analysis; L.A.W. and C.L.S. were involved with M. tuberculosis infection; E.G. and H.S. helped to identify EMSA probes; P.L., W.L. and W.J.L. performed ChIP-seq and helped to identify EMSA probes; M.B. and L.D.S. aided with T. gondii infections; S.S.K.L., C.T.A. and R.D.S. aided with tumour models. H.S. and W.J.L. provided helpful discussions. K.M.M. directed the work and wrote the manuscript. All authors discussed the results and contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-15, Supplementary Table 1 and additional references. (PDF 5221 kb)
Rights and permissions
About this article
Cite this article
Tussiwand, R., Lee, WL., Murphy, T. et al. Compensatory dendritic cell development mediated by BATF–IRF interactions. Nature 490, 502–507 (2012). https://doi.org/10.1038/nature11531
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11531
This article is cited by
-
CaSSiDI: novel single-cell “Cluster Similarity Scoring and Distinction Index” reveals critical functions for PirB and context-dependent Cebpb repression
Cell Death & Differentiation (2024)
-
siRNA-mediated downregulation of BATF3 diminished proliferation and induced apoptosis through downregulating c-Myc expression in chronic myelogenous leukemia cells
Molecular Biology Reports (2024)
-
Signaling pathways involved in the biological functions of dendritic cells and their implications for disease treatment
Molecular Biomedicine (2023)
-
Long non coding RNAs reveal important pathways in childhood asthma: a future perspective
Journal of Molecular Histology (2023)
-
Cisplatin resistance-related multi-omics differences and the establishment of machine learning models
Journal of Translational Medicine (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.