Abstract
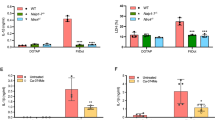
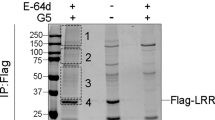
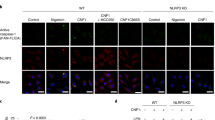
NLRC4 is a cytosolic member of the NOD-like receptor family that is expressed in innate immune cells. It senses indirectly bacterial flagellin and type III secretion systems, and responds by assembling an inflammasome complex that promotes caspase-1 activation and pyroptosis1,2,3,4,5,6. Here we use knock-in mice expressing NLRC4 with a carboxy-terminal 3×Flag tag to identify phosphorylation of NLRC4 on a single, evolutionarily conserved residue, Ser 533, following infection of macrophages with Salmonella enterica serovar Typhimurium (also known as Salmonella typhimurium). Western blotting with a NLRC4 phospho-Ser 533 antibody confirmed that this post-translational modification occurs only in the presence of stimuli known to engage NLRC4 and not the related protein NLRP3 or AIM2. Nlrc4−/− macrophages reconstituted with NLRC4 mutant S533A, unlike those reconstituted with wild-type NLRC4, did not activate caspase-1 and pyroptosis in response to S. typhimurium, indicating that S533 phosphorylation is critical for NLRC4 inflammasome function. Conversely, phosphomimetic NLRC4 S533D caused rapid macrophage pyroptosis without infection. Biochemical purification of the NLRC4-phosphorylating activity and a screen of kinase inhibitors identified PRKCD (PKCδ) as a candidate NLRC4 kinase. Recombinant PKCδ phosphorylated NLRC4 S533 in vitro, immunodepletion of PKCδ from macrophage lysates blocked NLRC4 S533 phosphorylation in vitro, and Prkcd−/− macrophages exhibited greatly attenuated caspase-1 activation and IL-1β secretion specifically in response to S. typhimurium. Phosphorylation-defective NLRC4 S533A failed to recruit procaspase-1 and did not assemble inflammasome specks7 during S. typhimurium infection, so phosphorylation of NLRC4 S533 probably drives conformational changes necessary for NLRC4 inflammasome activity and host innate immunity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mariathasan, S. et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218 (2004)
Miao, E. A. et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nature Immunol. 7, 569–575 (2006)
Franchi, L. et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in Salmonella-infected macrophages. Nature Immunol. 7, 576–582 (2006)
Miao, E. A. et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl Acad. Sci. USA 107, 3076–3080 (2010)
Kofoed, E. M. & Vance, R. E. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595 (2011)
Zhao, Y. et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596–600 (2011)
Broz, P. et al. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207, 1745–1755 (2010)
Amer, A. et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J. Biol. Chem. 281, 35217–35223 (2006)
Mariathasan, S. et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440, 228–232 (2006)
Rathinam, V. A. et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nature Immunol. 11, 395–402 (2010)
Fernandes-Alnemri, T. et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nature Immunol. 11, 385–393 (2010)
Jones, J. W. et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl Acad. Sci. USA 107, 9771–9776 (2010)
Hersh, D. et al. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl Acad. Sci. USA 96, 2396–2401 (1999)
Wang, G. G. et al. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nature Methods 3, 287–293 (2006)
Poyet, J. L. et al. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276, 28309–28313 (2001)
Wolf, M. & Baggiolini, M. The protein kinase inhibitor staurosporine, like phorbol esters, induces the association of protein kinase C with membranes. Biochem. Biophys. Res. Commun. 154, 1273–1279 (1988)
Mizuno, K., Saido, T. C., Ohno, S., Tamaoki, T. & Suzuki, K. Staurosporine-related compounds, K252a and UCN-01, inhibit both cPKC and nPKC. FEBS Lett. 330, 114–116 (1993)
Steinberg, S. F. Structural basis of protein kinase C isoform function. Physiol. Rev. 88, 1341–1378 (2008)
Soltoff, S. P. Rottlerin: an inappropriate and ineffective inhibitor of PKCδ. Trends Pharmacol. Sci. 28, 453–458 (2007)
Agostini, L. et al. NALP3 forms an IL-1β-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity 20, 319–325 (2004)
Vendel, A. C. et al. B and T lymphocyte attenuator regulates B cell receptor signaling by targeting Syk and BLNK. J. Immunol. 182, 1509–1517 (2009)
Castellana, N. E. et al. Resurrection of a clinical antibody: template proteogenomic de novo proteomic sequencing and reverse engineering of an anti-lymphotoxin-α antibody. Proteomics 11, 395–405 (2011)
Wang, G. G. et al. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nature Methods 3, 287–293 (2006)
Acknowledgements
We thank P. Broz, A. Paler Martinez, M. Roose-Girma, X. Rairdan, C. Kung, V. Asghari and S. Mukund for technical support, and K. O’Rourke for editorial assistance.
Author information
Authors and Affiliations
Contributions
Y.Q., S.M., A.I.-T., D.A., L.L.G., J.E.C., L.K. and S.L. designed and conducted experiments; K.N. generated the Nlrc4 F/F mice; J.L. performed bioinformatics analyses; M.L., N.K. and D.M. discussed the study; Y.Q., K.N. and V.M.D. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Y.Q., S.M., A.I.-T., K.N., L.L.G., J.E.C., S.L., N.K., J.L., L.K., D.A. and V.M.D. are employees of Genentech, Inc.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-6 and Supplementary Tables 1-2. (PDF 2399 kb)
Rights and permissions
About this article
Cite this article
Qu, Y., Misaghi, S., Izrael-Tomasevic, A. et al. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature 490, 539–542 (2012). https://doi.org/10.1038/nature11429
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11429
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.