Abstract
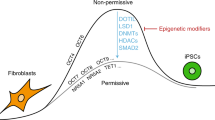
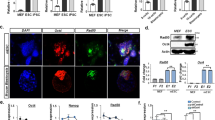
Somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) by using the pluripotency factors Oct4, Sox2, Klf4 and c-Myc (together referred to as OSKM)1. iPSC reprogramming erases somatic epigenetic signatures—as typified by DNA methylation or histone modification at silent pluripotency loci—and establishes alternative epigenetic marks of embryonic stem cells (ESCs)2. Here we describe an early and essential stage of somatic cell reprogramming, preceding the induction of transcription at endogenous pluripotency loci such as Nanog and Esrrb. By day 4 after transduction with OSKM, two epigenetic modification factors necessary for iPSC generation, namely poly(ADP-ribose) polymerase-1 (Parp1) and ten-eleven translocation-2 (Tet2), are recruited to the Nanog and Esrrb loci. These epigenetic modification factors seem to have complementary roles in the establishment of early epigenetic marks during somatic cell reprogramming: Parp1 functions in the regulation of 5-methylcytosine (5mC) modification, whereas Tet2 is essential for the early generation of 5-hydroxymethylcytosine (5hmC) by the oxidation of 5mC (refs 3,4). Although 5hmC has been proposed to serve primarily as an intermediate in 5mC demethylation to cytosine in certain contexts5,6,7, our data, and also studies of Tet2-mutant human tumour cells8, argue in favour of a role for 5hmC as an epigenetic mark distinct from 5mC. Consistent with this, Parp1 and Tet2 are each needed for the early establishment of histone modifications that typify an activated chromatin state at pluripotency loci, whereas Parp1 induction further promotes accessibility to the Oct4 reprogramming factor. These findings suggest that Parp1 and Tet2 contribute to an epigenetic program that directs subsequent transcriptional induction at pluripotency loci during somatic cell reprogramming.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006)
Mikkelsen, T. S. et al. Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 (2008)
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009)
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009)
Ito, S. et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 (2010)
Figueroa, M. E. et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567 (2010)
Guo, J. U. et al. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 (2011)
Ko, M. et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468, 839–843 (2011)
Krishnakumar, R. & Kraus, W. L. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell 39, 8–24 (2010)
Wacker, D. A. et al. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol. Cell. Biol. 27, 7475–7485 (2007)
Langelier, M. F. et al. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 285, 18877–18887 (2010)
Hajkova, P. et al. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329, 78–82 (2010)
Williams, K. et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 (2011)
Pastor, W. A. et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473, 394–397 (2011)
Wu, H. et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 25, 679–684 (2011)
Ficz, G. et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402 (2011)
Wu, H. et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473, 389–393 (2011)
Davis, T. & Vaisvila, R. High sensitivity 5-hydroxymethylcytosine detection in Balb/C brain tissue. J. Vis. Exp.. (48), e2661, http://dx.doi.org/10.3791/2661 (2011)
Meissner, A. et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770 (2008)
Bhutani, N., Burns, D. M. & Blau, H. M. DNA demethylation dynamics. Cell 146, 866–872 (2011)
Yildirim, O. et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147, 1498–1510 (2011)
Bernstein, B. E. et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181 (2005)
Heintzman, N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genet. 39, 311–318 (2007)
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006)
Lee, T. I. et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 (2006)
Mikkelsen, T. S. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007)
Koh, K. P. et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200–213 (2011)
Wang, Z. Q. et al. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 9, 509–520 (1995)
Moran-Crusio, K. et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20, 11–24 (2011)
Smith, Z. D., Nachman, I., Regev, A. & Meissner, A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nature Biotechnol. 28, 521–526 (2010)
Acknowledgements
We thank G. Q. Daley, A. P. Feinberg, A. Doi, R. M. Santella and M. A. Kappil for reagents and for technical assistance with pyrosequencing; A. Califano and A. Lachmann for assistance with the bioinformatics analyses; E. O. Mazzoni for assistance with the ChIP analyses; and O. Hobert for critical reading of the manuscript. This work was supported by New York State Stem Cell Science (NYSTEM) grants C024402 and C024403 and National Institutes of Health (NIH) grant RO1 NS064433 to A.A., NYSTEM Institution Development Grant N08G-071 to E.I.C., NIH grant RO1 138424 to R.L.L, and a shared NIH/National Center for Research Resources instrument grant for mass spectrometry, 1 S10 RR023680-1.
Author information
Authors and Affiliations
Contributions
C.A.D. and A.A. designed the experiments and analysed data. C.A.D., D.B.R., S.T., R.F. and W.B.V. conducted molecular and cellular experiments. T.Y., G.B. and K.I. performed and analysed murine in vivo studies. R.L.L. and A.S. supplied essential reagents. P.G. performed bioinformatics analyses. S.N. and E.I.C. conducted proteomics. C.A.D. and A.A. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-5, Supplementary Methods, a legend for Supplementary Table 1 (see separate excel file), Supplementary Tables 2-9 and Supplementary References. (PDF 1471 kb)
Supplementary Table 1
This file contains the mass spectrometry raw data – see legend in Supplementary Information file. (XLS 246 kb)
Rights and permissions
About this article
Cite this article
Doege, C., Inoue, K., Yamashita, T. et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488, 652–655 (2012). https://doi.org/10.1038/nature11333
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11333
This article is cited by
-
Genome-wide ATAC-see screening identifies TFDP1 as a modulator of global chromatin accessibility
Nature Genetics (2024)
-
TET2 is required to suppress mTORC1 signaling through urea cycle with therapeutic potential
Cell Discovery (2023)
-
Role of NAD+ and FAD in Ischemic Stroke Pathophysiology: An Epigenetic Nexus and Expanding Therapeutic Repertoire
Cellular and Molecular Neurobiology (2023)
-
Sin3a drives mesenchymal-to-epithelial transition through cooperating with Tet1 in somatic cell reprogramming
Stem Cell Research & Therapy (2022)
-
AKT signaling is associated with epigenetic reprogramming via the upregulation of TET and its cofactor, alpha-ketoglutarate during iPSC generation
Stem Cell Research & Therapy (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.