Abstract
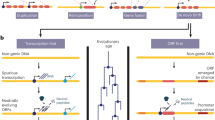
Novel protein-coding genes can arise either through re-organization of pre-existing genes or de novo1,2. Processes involving re-organization of pre-existing genes, notably after gene duplication, have been extensively described1,2. In contrast, de novo gene birth remains poorly understood, mainly because translation of sequences devoid of genes, or ‘non-genic’ sequences, is expected to produce insignificant polypeptides rather than proteins with specific biological functions1,3,4,5,6. Here we formalize an evolutionary model according to which functional genes evolve de novo through transitory proto-genes4 generated by widespread translational activity in non-genic sequences. Testing this model at the genome scale in Saccharomyces cerevisiae, we detect translation of hundreds of short species-specific open reading frames (ORFs) located in non-genic sequences. These translation events seem to provide adaptive potential7, as suggested by their differential regulation upon stress and by signatures of retention by natural selection. In line with our model, we establish that S. cerevisiae ORFs can be placed within an evolutionary continuum ranging from non-genic sequences to genes. We identify ∼1,900 candidate proto-genes among S. cerevisiae ORFs and find that de novo gene birth from such a reservoir may be more prevalent than sporadic gene duplication. Our work illustrates that evolution exploits seemingly dispensable sequences to generate adaptive functional innovation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tautz, D. & Domazet-Loso, T. The evolutionary origin of orphan genes. Nature Rev. Genet. 12, 692–702 (2011)
Kaessmann, H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 20, 1313–1326 (2010)
Jacob, F. Evolution and tinkering. Science 196, 1161–1166 (1977)
Siepel, A. Darwinian alchemy: human genes from noncoding DNA. Genome Res. 19, 1693–1695 (2009)
Khalturin, K., Hemmrich, G., Fraune, S., Augustin, R. & Bosch, T. C. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 25, 404–413 (2009)
Wilson, B. A. & Masel, J. Putatively noncoding transcripts show extensive association with ribosomes. Genome Biol. Evol. 3, 1245–1252 (2011)
Jarosz, D. F., Taipale, M. & Lindquist, S. Protein homeostasis and the phenotypic manifestation of genetic diversity: principles and mechanisms. Annu. Rev. Genet. 44, 189–216 (2010)
Cai, J., Zhao, R., Jiang, H. & Wang, W. De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics 179, 487–496 (2008)
Wu, D. D., Irwin, D. M. & Zhang, Y. P. De novo origin of human protein-coding genes. PLoS Genet. 7, e1002379 (2011)
Ekman, D. & Elofsson, A. Identifying and quantifying orphan protein sequences in fungi. J. Mol. Biol. 396, 396–405 (2010)
Lipman, D. J., Souvorov, A., Koonin, E. V., Panchenko, A. R. & Tatusova, T. A. The relationship of protein conservation and sequence length. BMC Evol. Biol. 2, 20 (2002)
Wolf, Y. I., Novichkov, P. S., Karev, G. P., Koonin, E. V. & Lipman, D. J. The universal distribution of evolutionary rates of genes and distinct characteristics of eukaryotic genes of different apparent ages. Proc. Natl Acad. Sci. USA 106, 7273–7280 (2009)
Cai, J. J. & Petrov, D. A. Relaxed purifying selection and possibly high rate of adaptation in primate lineage-specific genes. Genome Biol. Evol. 2, 393–409 (2010)
Zheng, D. & Gerstein, M. B. The ambiguous boundary between genes and pseudogenes: the dead rise up, or do they? Trends Genet. 23, 219–224 (2007)
Oliver, S. G. et al. The complete DNA sequence of yeast chromosome III. Nature 357, 38–46 (1992)
Fisk, D. G. et al. Saccharomyces cerevisiae S288C genome annotation: a working hypothesis. Yeast 23, 857–865 (2006)
Nagalakshmi, U. et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 (2008)
Boyer, J. et al. Large-scale exploration of growth inhibition caused by overexpression of genomic fragments in Saccharomyces cerevisiae. Genome Biol. 5, R72 (2004)
Brar, G. A. et al. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335, 552–557 (2012)
Li, Q. R. et al. Revisiting the Saccharomyces cerevisiae predicted ORFeome. Genome Res. 18, 1294–1303 (2008)
Jansen, R. & Gerstein, M. Analysis of the yeast transcriptome with structural and functional categories: characterizing highly expressed proteins. Nucleic Acids Res. 28, 1481–1488 (2000)
Giacomelli, M. G., Hancock, A. S. & Masel, J. The conversion of 3′ UTRs into coding regions. Mol. Biol. Evol. 24, 457–464 (2007)
Prat, Y., Fromer, M., Linial, N. & Linial, M. Codon usage is associated with the evolutionary age of genes in metazoan genomes. BMC Evol. Biol. 9, 285 (2009)
Yomtovian, I., Teerakulkittipong, N., Lee, B., Moult, J. & Unger, R. Composition bias and the origin of ORFan genes. Bioinformatics 26, 996–999 (2010)
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009)
Vishnoi, A., Kryazhimskiy, S., Bazykin, G. A., Hannenhalli, S. & Plotkin, J. B. Young proteins experience more variable selection pressures than old proteins. Genome Res. 20, 1574–1581 (2010)
Gao, L. Z. & Innan, H. Very low gene duplication rate in the yeast genome. Science 306, 1367–1370 (2004)
Hayden, E. J., Ferrada, E. & Wagner, A. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature 474, 92–95 (2011)
Pal, C., Papp, B. & Hurst, L. D. Highly expressed genes in yeast evolve slowly. Genetics 158, 927–931 (2001)
Drummond, D. A., Raval, A. & Wilke, C. O. A single determinant dominates the rate of yeast protein evolution. Mol. Biol. Evol. 23, 327–337 (2006)
Acknowledgements
We thank L. Duret, E. Levy, J. Vandenhaute, Q. Li, H. Yu, P. Braun, M. Dreze, C. Foo, M. Mann, N. Kulak, J. Cox, C. Maire and S. Jhavery-Schneider as well as members of the Center for Cancer Systems Biology (CCSB), in particular A. Dricot-Ziter, A. MacWilliams, F. Roth, Y. Jacob and D. Hill for discussions and proofreading. A.R. was supported by a National Institute of Health Pioneer Award, a Career Award at the Scientific Interface from the Burroughs Wellcome Fund and the Howard Hughes Medical Institute (HHMI). I.W. is a HHMI fellow of the Damon Runyon Cancer Research Institute. G.A.B. was supported by American Cancer Society Postdoctoral fellowship 117945-PF-09-136-01-RMC. M.V. is a Chercheur Qualifié Honoraire from the Fonds de la Recherche Scientifique (FRS-FNRS, Wallonia-Brussels Federation, Belgium). This work was supported by the grant R01-HG006061 from the National Human Genome Research Institute awarded to M.V.
Author information
Authors and Affiliations
Contributions
A.-R.C., I.W., M.E.C. and M.V. conceived the project. A.-R.C. led the project and performed most of the analyses. T.R. evaluated cross-species transfer events, optimized the ribosome footprint analysis pipeline and assisted in other analyses. I.W. designed the conservation level tool and calculated most of the purifying selection statistics. M.A.C., C.A.H., A.R. and N.T.-M. advised on the research. M.A.Y. aligned the sequencing reads. B.S. predicted disordered and transmembrane regions and assisted in the cross-species transfer analyses. N.S. and B.C. assisted in analyses. G.A.B. and J.S.W. shared their expertise in ribosome footprinting data analysis and provided the meiosis ribosome footprinting raw and processed data. A.-R.C., T.R., M.E.C. and M.V. designed the figures. All authors contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-8, Supplementary Methods, Supplementary Tables 1-4 and additional references. (PDF 11377 kb)
Rights and permissions
About this article
Cite this article
Carvunis, AR., Rolland, T., Wapinski, I. et al. Proto-genes and de novo gene birth. Nature 487, 370–374 (2012). https://doi.org/10.1038/nature11184
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11184
This article is cited by
-
The origin and structural evolution of de novo genes in Drosophila
Nature Communications (2024)
-
Small open reading frames: a comparative genetics approach to validation
BMC Genomics (2023)
-
Uncovering gene-family founder events during major evolutionary transitions in animals, plants and fungi using GenEra
Genome Biology (2023)
-
De novo gene increases brain size
Nature Ecology & Evolution (2023)
-
De novo genes with an lncRNA origin encode unique human brain developmental functionality
Nature Ecology & Evolution (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.