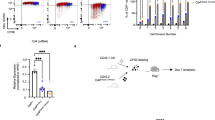
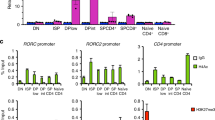
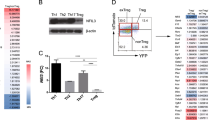
Abstract
During immune responses, naive CD4+ T cells differentiate into several T helper (TH) cell subsets under the control of lineage-specifying genes. These subsets (TH1, TH2 and TH17 cells and regulatory T cells) secrete distinct cytokines and are involved in protection against different types of infection. Epigenetic mechanisms are involved in the regulation of these developmental programs, and correlations have been drawn between the levels of particular epigenetic marks and the activity or silencing of specifying genes during differentiation1,2,3. Nevertheless, the functional relevance of the epigenetic pathways involved in TH cell subset differentiation and commitment is still unclear. Here we explore the role of the SUV39H1–H3K9me3–HP1α silencing pathway in the control of TH2 lineage stability. This pathway involves the histone methylase SUV39H1, which participates in the trimethylation of histone H3 on lysine 9 (H3K9me3), a modification that provides binding sites for heterochromatin protein 1α (HP1α)4,5 and promotes transcriptional silencing. This pathway was initially associated with heterochromatin formation and maintenance6 but can also contribute to the regulation of euchromatic genes7,8,9. We now propose that the SUV39H1–H3K9me3–HP1α pathway participates in maintaining the silencing of TH1 loci, ensuring TH2 lineage stability. In TH2 cells that are deficient in SUV39H1, the ratio between trimethylated and acetylated H3K9 is impaired, and the binding of HP1α at the promoters of silenced TH1 genes is reduced. Despite showing normal differentiation, both SUV39H1-deficient TH2 cells and HP1α-deficient TH2 cells, in contrast to wild-type cells, expressed TH1 genes when recultured under conditions that drive differentiation into TH1 cells. In a mouse model of TH2-driven allergic asthma, the chemical inhibition or loss of SUV39H1 skewed T-cell responses towards TH1 responses and decreased the lung pathology. These results establish a link between the SUV39H1–H3K9me3–HP1α pathway and the stability of TH2 cells, and they identify potential targets for therapeutic intervention in TH2-cell-mediated inflammatory diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Avni, O. et al. TH cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nature Immunol. 3, 643–651 (2002)
Wei, G. et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167 (2009)
Chang, S. & Aune, T. M. Dynamic changes in histone-methylation ‘marks’ across the locus encoding interferon-γ during the differentiation of T helper type 2 cells. Nature Immunol. 8, 723–731 (2007)
Lachner, M., O’Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001)
Bannister, A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001)
Maison, C. & Almouzni, G. HP1 and the dynamics of heterochromatin maintenance. Nature Rev. Mol. Cell Biol. 5, 296–305 (2004)
Nielsen, S. J. et al. Rb targets histone H3 methylation and HP1 to promoters. Nature 412, 561–565 (2001)
Wiencke, J. K., Zheng, S., Morrison, Z. & Yeh, R. F. Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells. Oncogene 27, 2412–2421 (2008)
Ait-Si-Ali, S. et al. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 23, 605–615 (2004)
Wilson, C. B., Rowell, E. & Sekimata, M. Epigenetic control of T-helper-cell differentiation. Nature Rev. Immunol. 9, 91–105 (2009)
Peters, A. H. et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001)
Grogan, J. L. et al. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity 14, 205–215 (2001)
Wang, H. et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell 12, 475–487 (2003)
Balasubramani, A., Mukasa, R., Hatton, R. D. & Weaver, C. T. Regulation of the Ifng locus in the context of T-lineage specification and plasticity. Immunol. Rev. 238, 216–232 (2010)
Vaute, O., Nicolas, E., Vandel, L. & Trouche, D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 30, 475–481 (2002)
Lighvani, A. A. et al. T-bet is rapidly induced by interferon-γ in lymphoid and myeloid cells. Proc. Natl Acad. Sci. USA 98, 15137–15142 (2001)
Afkarian, M. et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nature Immunol. 3, 549–557 (2002)
Yamamoto, K. & Sonoda, M. Self-interaction of heterochromatin protein 1 is required for direct binding to histone methyltransferase, SUV39H1. Biochem. Biophys. Res. Commun. 301, 287–292 (2003)
Aagaard, L. et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 18, 1923–1938 (1999)
Eskeland, R., Eberharter, A. & Imhof, A. HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Mol. Cell. Biol. 27, 453–465 (2007)
Mateescu, B., England, P., Halgand, F., Yaniv, M. & Muchardt, C. Tethering of HP1 proteins to chromatin is relieved by phosphoacetylation of histone H3. EMBO Rep. 5, 490–496 (2004)
Kumar, R. K., Herbert, C. & Foster, P. S. The ‘classical’ ovalbumin challenge model of asthma in mice. Curr. Drug Targets 9, 485–494 (2008)
Snapper, C. M., Peschel, C. & Paul, W. E. IFN-γ stimulates IgG2a secretion by murine B cells stimulated with bacterial lipopolysaccharide. J. Immunol. 140, 2121–2127 (1988)
Gonzalo, J. A. et al. Eosinophil recruitment to the lung in a murine model of allergic inflammation. The role of T cells, chemokines, and adhesion receptors. J. Clin. Invest. 98, 2332–2345 (1996)
Romagnani, S. Immunologic influences on allergy and the TH1/TH2 balance. J. Allergy Clin. Immunol. 113, 395–400 (2004)
Barnes, P. J. The cytokine network in asthma and chronic obstructive pulmonary disease. J. Clin. Invest. 118, 3546–3556 (2008)
Barnes, P. J. Immunology of asthma and chronic obstructive pulmonary disease. Nature Rev. Immunol. 8, 183–192 (2008)
Greiner, D., Bonaldi, T., Eskeland, R., Roemer, E. & Imhof, A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nature Chem. Biol. 1, 143–145 (2005)
Lee, T. I., Johnstone, S. E. & Young, R. A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nature Protocols 1, 729–748 (2006)
Acknowledgements
We are grateful to the late R. Losson for the generation of the HP1α-deficient mice. We acknowledge the members of the Department of Pathology and the Nikon Imaging Centre at the Institut Curie-CNRS. We thank T. Jenuwein for providing the SUV39H1-deficient mice and the members of INSERM U932 and CNRS UMR218 for discussions and suggestions. This work was supported by ANR-09-BLAN-0257 (‘ECens’), ANR 2010 1326 03 and HEALTH-F4-2010-257082 (from the European Commission Network of Excellence EpiGeneSys) to G.A., ANR 2009 BLAN-0021 EPIGO to F.C., and ANR 2010 BLAN-1326 01 and a grant from the Ligue National de Lutte contre le Cancer (Ligue équipe labélisée 2011–2013) to S.A. and E.Z. E.Z. and H.A.S. were funded by Fellowship of the Institut Curie (Paris), and R.S.A. was funded by an Australian National Health and Medical Research Council-INSERM fellowship (461286).
Author information
Authors and Affiliations
Contributions
R.S.A., E.Z. and F.C. are joint first authors. R.S.A. and E.Z. designed the project, carried out experimental work and wrote the manuscript. F.C. carried out experimental work and interpreted data. H.A.S. carried out experimental work, analysed data and participated in data interpretation. V.M. and D.R. carried out experimental work. G.T.B. provided critical materials. C.M. and J.-P.Q. provided critical materials and wrote the manuscript. G.A. supervised the research and wrote the manuscript. S.A. conceived the project, supervised the research and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-10 and Supplementary Table 1 with additional references. (PDF 9820 kb)
Rights and permissions
About this article
Cite this article
Allan, R., Zueva, E., Cammas, F. et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature 487, 249–253 (2012). https://doi.org/10.1038/nature11173
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11173
This article is cited by
-
Epigenetic regulation in the tumor microenvironment: molecular mechanisms and therapeutic targets
Signal Transduction and Targeted Therapy (2023)
-
CRL4B complex-mediated H2AK119 monoubiquitination restrains Th1 and Th2 cell differentiation
Cell Death & Differentiation (2023)
-
Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance
Nature Reviews Molecular Cell Biology (2022)
-
CD8+T cell responsiveness to anti-PD-1 is epigenetically regulated by Suv39h1 in melanomas
Nature Communications (2022)
-
Suppressor of variegation 3–9 homologue 1 impairment and neutrophil-skewed systemic inflammation are associated with comorbidities in COPD
BMC Pulmonary Medicine (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.