Abstract
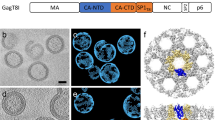
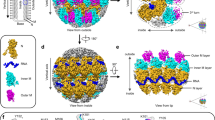
The assembly of retroviruses such as HIV-1 is driven by oligomerization of their major structural protein, Gag. Gag is a multidomain polyprotein including three conserved folded domains: MA (matrix), CA (capsid) and NC (nucleocapsid)1. Assembly of an infectious virion proceeds in two stages2. In the first stage, Gag oligomerization into a hexameric protein lattice leads to the formation of an incomplete, roughly spherical protein shell that buds through the plasma membrane of the infected cell to release an enveloped immature virus particle. In the second stage, cleavage of Gag by the viral protease leads to rearrangement of the particle interior, converting the non-infectious immature virus particle into a mature infectious virion. The immature Gag shell acts as the pivotal intermediate in assembly and is a potential target for anti-retroviral drugs both in inhibiting virus assembly and in disrupting virus maturation3. However, detailed structural information on the immature Gag shell has not previously been available. For this reason it is unclear what protein conformations and interfaces mediate the interactions between domains and therefore the assembly of retrovirus particles, and what structural transitions are associated with retrovirus maturation. Here we solve the structure of the immature retroviral Gag shell from Mason–Pfizer monkey virus by combining cryo-electron microscopy and tomography. The 8-Å resolution structure permits the derivation of a pseudo-atomic model of CA in the immature retrovirus, which defines the protein interfaces mediating retrovirus assembly. We show that transition of an immature retrovirus into its mature infectious form involves marked rotations and translations of CA domains, that the roles of the amino-terminal and carboxy-terminal domains of CA in assembling the immature and mature hexameric lattices are exchanged, and that the CA interactions that stabilize the immature and mature viruses are almost completely distinct.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Göttlinger, H. G. The HIV-1 assembly machine. AIDS 15 (Suppl. 5). S13–S20 (2001)
Briggs, J. A. & Kräusslich, H. G. The molecular architecture of HIV. J. Mol. Biol. 410, 491–500 (2011)
Waheed, A. A. & Freed, E. O. HIV type 1 Gag as a target for antiviral therapy. AIDS Res. Hum. Retroviruses 28, 54–75 (2012)
Gross, I., Hohenberg, H., Huckhagel, C. & Kräusslich, H. G. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72, 4798–4810 (1998)
von Schwedler, U. K. et al. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17, 1555–1568 (1998)
Johnson, M. C., Scobie, H. M., Ma, Y. M. & Vogt, V. M. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76, 11177–11185 (2002)
Accola, M. A., Strack, B. & Göttlinger, H. G. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74, 5395–5402 (2000)
Yeager, M., Wilson-Kubalek, E. M., Weiner, S. G., Brown, P. O. & Rein, A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl Acad. Sci. USA 95, 7299–7304 (1998)
Li, S., Hill, C. P., Sundquist, W. I. & Finch, J. T. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407, 409–413 (2000)
Briggs, J. A., Wilk, T., Welker, R., Kräusslich, H. G. & Fuller, S. D. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22, 1707–1715 (2003)
Briggs, J. A. et al. The stoichiometry of Gag protein in HIV-1. Nature Struct. Mol. Biol. 11, 672–675 (2004)
Ganser-Pornillos, B. K., Cheng, A. & Yeager, M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell 131, 70–79 (2007)
Pornillos, O. et al. X-ray structures of the hexameric building block of the HIV capsid. Cell 137, 1282–1292 (2009)
Pornillos, O., Ganser-Pornillos, B. K. & Yeager, M. Atomic-level modelling of the HIV capsid. Nature 469, 424–427 (2011)
Cardone, G., Purdy, J. G., Cheng, N., Craven, R. C. & Steven, A. C. Visualization of a missing link in retrovirus capsid assembly. Nature 457, 694–698 (2009)
Briggs, J. A. et al. Structure and assembly of immature HIV. Proc. Natl Acad. Sci. USA 106, 11090–11095 (2009)
Wright, E. R. et al. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26, 2218–2226 (2007)
de Marco, A. et al. Conserved and variable features of Gag structure and arrangement in immature retrovirus particles. J. Virol. 84, 11729–11736 (2010)
Ulbrich, P. et al. Distinct roles for nucleic acid in in vitro assembly of purified Mason-Pfizer monkey virus CANC proteins. J. Virol. 80, 7089–7099 (2006)
Sachse, C. et al. High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus. J. Mol. Biol. 371, 812–835 (2007)
Egelman, E. H. Reconstruction of helical filaments and tubes. Methods Enzymol. 482, 167–183 (2010)
Macek, P. et al. NMR structure of the N-terminal domain of capsid protein from the Mason-Pfizer monkey virus. J. Mol. Biol. 392, 100–114 (2009)
de Marco, A. et al. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog. 6, e1001215 (2010)
Lanman, J. et al. Key interactions in HIV-1 maturation identified by hydrogen–deuterium exchange. Nature Struct. Mol. Biol. 11, 676–677 (2004)
Ternois, F., Sticht, J., Duquerroy, S., Kräusslich, H. G. & Rey, F. A. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nature Struct. Mol. Biol. 12, 678–682 (2005)
Bartonova, V. et al. Residues in the HIV-1 capsid assembly inhibitor binding site are essential for maintaining the assembly-competent quaternary structure of the capsid protein. J. Biol. Chem. 283, 32024–32033 (2008)
Chu, H. H., Chang, Y. F. & Wang, C. T. Mutations in the alpha-helix directly C-terminal to the major homology region of human immunodeficiency virus type 1 capsid protein disrupt Gag multimerization and markedly impair virus particle production. J. Biomed. Sci. 13, 645–656 (2006)
von Schwedler, U. K., Stray, K. M., Garrus, J. E. & Sundquist, W. I. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77, 5439–5450 (2003)
Yu, I. M. et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319, 1834–1837 (2008)
Conway, J. F. et al. Virus maturation involving large subunit rotations and local refolding. Science 292, 744–748 (2001)
Acknowledgements
This study was technically supported by the use of the European Molecular Biology Laboratory Information Technology Service unit. This work was partly funded by a grant from the Deutsche Forschungsgemeinschaft within SPP1175 to J.A.G.B. and by grants P302/12/1895 and 204/09/1388 from the Czech Science foundation to T.R. and M.R.
Author information
Authors and Affiliations
Contributions
T.A.M.B., P.U., M.R., T.R. and J.A.G.B. designed the research. T.A.M.B. and P.U. prepared samples for electron microscopy. T.A.M.B. and J.D.R. collected cryo-EM data. T.A.M.B., J.D.R., A.D.M. and J.A.G.B. analysed cryo-ET data. C.S. supported helical image-processing techniques. T.A.M.B. and J.A.G.B. developed and applied the variable-symmetry helical reconstruction methodology. T.A.M.B., N.D., P.U., M.R., C.S., T.R. and J.A.G.B. analysed fitted pseudo-atomic models. T.A.M.B. and J.A.G.B. wrote the paper with support from all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-8, Supplementary Methods, Supplementary Tables 1-2, legends for Supplementary Movies 1-2 and Supplementary References. (PDF 3688 kb)
Supplementary Movie 1
In this movie we see the structure of the M-PMV CANC tubes at sub-nanometre resolution - see Supplementary Information file for full legend. (MOV 16313 kb)
Supplementary Movie 2
In this movie we see the comparison of the immature and mature retroviral lattices - see Supplementary Information file for full legend. (MOV 5283 kb)
Rights and permissions
About this article
Cite this article
Bharat, T., Davey, N., Ulbrich, P. et al. Structure of the immature retroviral capsid at 8 Å resolution by cryo-electron microscopy. Nature 487, 385–389 (2012). https://doi.org/10.1038/nature11169
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11169
This article is cited by
-
A single G10T polymorphism in HIV-1 subtype C Gag-SP1 regulates sensitivity to maturation inhibitors
Retrovirology (2021)
-
Recent advances in retroviruses via cryo-electron microscopy
Retrovirology (2018)
-
Expression, purification, and characterization of biologically active full-length Mason-Pfizer monkey virus (MPMV) Pr78Gag
Scientific Reports (2018)
-
Insights into the activity of maturation inhibitor PF-46396 on HIV-1 clade C
Scientific Reports (2017)
-
Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION
Nature Protocols (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.