Abstract
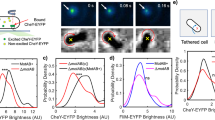
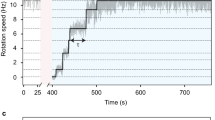
In the bacterial chemotaxis network, receptor clusters process input1,2,3, and flagellar motors generate output4. Receptor and motor complexes are coupled by the diffusible protein CheY-P. Receptor output (the steady-state concentration of CheY-P) varies from cell to cell5. However, the motor is ultrasensitive, with a narrow operating range of CheY-P concentrations6. How the match between receptor output and motor input might be optimized is unclear. Here we show that the motor can shift its operating range by changing its composition. The number of FliM subunits in the C-ring increases in response to a decrement in the concentration of CheY-P, increasing motor sensitivity. This shift in sensitivity explains the slow partial adaptation observed in mutants that lack the receptor methyltransferase and methylesterase7,8 and why motors show signal-dependent FliM turnover9. Adaptive remodelling is likely to be a common feature in the operation of many molecular machines.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sourjik, V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 12, 569–576 (2004)
Hazelbauer, G. L., Falke, J. J. & Parkinson, J. S. Baterial chemoreceptors: high -performance signaling in networked arrays. Trends Biochem. Sci. 33, 9–19 (2008)
Greenfield, D. et al. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 7, e1000137 (2009)
Berg, H. C. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72, 19–54 (2003)
Alon, U., Surette, M. G., Barkai, N. & Leibler, S. Robustness in bacterial chemotaxis. Nature 397, 168–171 (1999)
Cluzel, P., Surette, M. & Leibler, S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science 287, 1652–1655 (2000)
Stock, J., Kersulis, G. & Koshland, D. E., Jr Neither methylating nor demthylating enzymes are required for bacterial chemotaxis. Cell 42, 683–690 (1985)
Segall, J. E., Block, S. M. & Berg, H. C. Temporal comparisons in bacterial chemotaxis. Proc. Natl Acad. Sci. USA 83, 8987–8991 (1986)
Delalez, N. J. et al. Signal-dependent turnover of the bacterial flagellar switch protein FliM. Proc. Natl Acad. Sci. USA 107, 11347–11351 (2010)
Duke, T. A. J. & Bray, D. Heightened sensitivity of a lattice of membrane of receptors. Proc. Natl Acad. Sci. USA 96, 10104–10108 (1999)
Sourjik, V. & Berg, H. C. Receptor sensitivity in bacterial chemotaxis. Proc. Natl Acad. Sci. USA 99, 123–127 (2002)
Mello, B. A. & Tu, Y. Effects of adaptation in maintaining high sensitivity over a wide dynamic range of backgrounds for Escherichia coli chemotaxis. Biophys. J. 92, 2329–2337 (2007)
Hansen, C. H., Endres, R. G. & Wingreen, N. S. Chemotaxis in Escherichia coli: a molecular model for robust precise adaptation. PLoS Comput. Biol. 4, e1 (2008)
Levin, M. D., Morton-Firth, C. J., Abouhamad, W. N., Bourret, R. B. & Bray, D. Origins of individual swimming behavior in bacteria. Biophys. J. 74, 175–181 (1998)
Sourjik, V., Vaknin, A., Shimizu, T. S. & Berg, H. C. In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol. 423, 365–391 (2007)
Shimizu, T. S., Delalez, N., Pichler, K. & Berg, H. C. Monitoring bacterial chemotaxis by using bioluminescence resonance energy transfer: absence of feedback from the flagellar motors. Proc. Natl Acad. Sci. USA 103, 2093–2097 (2006)
Blair, D. F. & Berg, H. C. Restoration of torque in defective flagellar motors. Science 242, 1678–1681 (1988)
Reid, S. W. et al. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl Acad. Sci. USA 103, 8066–8071 (2006)
Leake, M. C. et al. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443, 355–358 (2006)
Fukuoka, H., Inoue, Y., Terasawa, S., Takahashi, H. & Ishijima, A. Exchange of rotor components in functioning bacterial flagellar motor. Biochem. Biophys. Res. Commun. 394, 130–135 (2010)
Yuan, J. & Berg, H. C. Resurrection of the flagellar motor near zero load. Proc. Natl Acad. Sci. USA 105, 1182–1185 (2008)
Lipkow, K. Changing cellular localization of CheZ predicted by molecular simulations. PLoS Comput. Biol. 2, e39 (2006)
Blat, Y. & Eisenbach, M. Oligomerization of the phosphatase CheZ upon interaction with the phosphorylated form of CheY, the signal protein of bacterial chemotaxis. J. Biol. Chem. 271, 1226–1231 (1996)
Sourjik, V. & Berg, H. C. Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA 99, 12669–12674 (2002)
Monod, J., Wyman, J. & Changeux, J. P. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12, 88–118 (1965)
Alon, U. et al. Response regulator output in bacterial chemotaxis. EMBO J. 17, 4238–4248 (1998)
Turner, L., Caplan, S. R. & Berg, H. C. Temperature induced switching of the bacterial flagellar motor. Biophys. J. 71, 2227–2233 (1996)
Sourjik, V. & Berg, H. C. Functional interactions between receptors in bacterial chemotaxis. Nature 428, 437–441 (2004)
Yuan, J., Fahrner, K. A. & Berg, H. C. Switching of the bacterial flagellar motor near zero load. J. Mol. Biol. 390, 394–400 (2009)
Shimizu, T. S., Tu, Y. & Berg, H. C. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Mol. Syst. Biol. 6, 382 (2010)
Parkinson, J. S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J. Bacteriol. 135, 45–53 (1978)
Amann, E., Ochs, B. & Abel, K.-J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69, 301–315 (1988)
Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 (1995)
Yuan, J., Fahrner, K. A., Turner, L. & Berg, H. C. Asymmetry in the clockwise and counter-clockwise rotation of the bacterial flagellar motor. Proc. Natl Acad. Sci. USA 107, 12846–12849 (2010)
Berg, H. C. & Block, S. M. A miniature flow cell designed for rapid exchange of media under high-power microscope objectives. J. Gen. Microbiol. 130, 2915–2920 (1984)
Thompson, R. E., Larson, D. R. & Webb, W. W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 82, 2775–2783 (2002)
Acknowledgements
This work was supported by National Institutes of Health Grant AI016478. R.W.B. is a recipient of an EMBO Long-Term Fellowship.
Author information
Authors and Affiliations
Contributions
J.Y. and H.C.B. planned the work and wrote the first draft of the paper. J.Y. performed the research with help on the MWC model and TIRF experiment and analysis from R.W.B. and on TIRF analysis from B.G.H.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Yuan, J., Branch, R., Hosu, B. et al. Adaptation at the output of the chemotaxis signalling pathway. Nature 484, 233–236 (2012). https://doi.org/10.1038/nature10964
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10964
This article is cited by
-
Bacterial motility: machinery and mechanisms
Nature Reviews Microbiology (2022)
-
Mechanosensitive recruitment of stator units promotes binding of the response regulator CheY-P to the flagellar motor
Nature Communications (2021)
-
Maximal information transmission is compatible with ultrasensitive biological pathways
Scientific Reports (2019)
-
Torque, but not FliL, regulates mechanosensitive flagellar motor-function
Scientific Reports (2017)
-
Impact of fluorescent protein fusions on the bacterial flagellar motor
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.