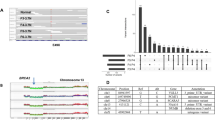
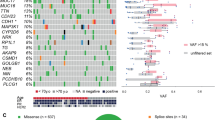
Abstract
Primary triple-negative breast cancers (TNBCs), a tumour type defined by lack of oestrogen receptor, progesterone receptor and ERBB2 gene amplification, represent approximately 16% of all breast cancers1. Here we show in 104 TNBC cases that at the time of diagnosis these cancers exhibit a wide and continuous spectrum of genomic evolution, with some having only a handful of coding somatic aberrations in a few pathways, whereas others contain hundreds of coding somatic mutations. High-throughput RNA sequencing (RNA-seq) revealed that only approximately 36% of mutations are expressed. Using deep re-sequencing measurements of allelic abundance for 2,414 somatic mutations, we determine for the first time—to our knowledge—in an epithelial tumour subtype, the relative abundance of clonal frequencies among cases representative of the population. We show that TNBCs vary widely in their clonal frequencies at the time of diagnosis, with the basal subtype of TNBC2,3 showing more variation than non-basal TNBC. Although p53 (also known as TP53), PIK3CA and PTEN somatic mutations seem to be clonally dominant compared to other genes, in some tumours their clonal frequencies are incompatible with founder status. Mutations in cytoskeletal, cell shape and motility proteins occurred at lower clonal frequencies, suggesting that they occurred later during tumour progression. Taken together, our results show that understanding the biology and therapeutic responses of patients with TNBC will require the determination of individual tumour clonal genotypes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Sequence Read Archive
Data deposits
Aligned exome/genome sequence data, RNA-seq data and Affymetrix SNP6.0 data sets are available at the European Genome-phenome Archive (http://www.ebi.ac.uk/ega/) under study accession number EGAS00001000132. Normal reference RNA-seq datasets are available at the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces) under study accession number SRP000930.
References
Blows, F. M. et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 7, e1000279 (2010)
Perou, C. M. et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000)
Sørlie, T. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA 98, 10869–10874 (2001)
Roth, A. et al. JointSNVmix: A probabilistic model for accurate detection of somatic mutations in normal/tumour paired next generation sequencing data. Bioinformatics http://dx.doi.org/10.1093/bioinformatics/bts053 (27 January 2012)
Ding, J. et al. Feature-based classifiers for somatic mutation detection in tumour—normal paired sequencing data. Bioinformatics 28, 167–175 (2012)
Ding, L. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075 (2008)
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature http://dx.doi.org/10.1038/nature10983 (this issue)
Poulogiannis, G. et al. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc. Natl Acad. Sci. USA 107, 15145–15150 (2010)
Bignell, G. R. et al. Signatures of mutation and selection in the cancer genome. Nature 463, 893–898 (2010)
Stephens, P. J. et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462, 1005–1010 (2009)
Morin, R. D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011)
Chicas, A. et al. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17, 376–387 (2010)
Herschkowitz, J. I., He, X., Fan, C. & Perou, C. M. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 10, R75 (2008)
Langerød, A. et al. TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res. 9, R30 (2007)
Børresen-Dale, A.-L. TP53 and breast cancer. Hum. Mutat. 21, 292–300 (2003)
Parker, J. S. et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167 (2009)
Youn, A. & Simon, R. Identifying cancer driver genes in tumor genome sequencing studies. Bioinformatics 27, 175–181 (2011)
Agrawal, N. et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333, 1154–1157 (2011)
Stransky, N. et al. The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160 (2011)
Wu, G., Feng, X. & Stein, L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 11, R53 (2010)
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011)
Shah, S. P. et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461, 809–813 (2009)
Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010)
Acknowledgements
The support of the BC Cancer Agency Tumour Bank, CBCF Breast Tumour Bank Alberta and the Addenbrookes Tumour bank (supported by NIHR and ECMC) is acknowledged. Technical support is acknowledged from the Centre for Translational Genomics, the Michael Smith Genome Sciences Centre technical group, the BCCA Flow Cytometry Core Facility in the Terry Fox Laboratory and the Cancer Research UK Cambridge Research Institute. Supported by the BC Cancer Foundation, US Department of Defense CDMRP program, Canadian Breast Cancer Foundation (BC Yukon) (to S.A. and S.S.), Michael Smith Foundation for Health Research (to S.S.), US National Institutes of Health (NIH) Roadmap Epigenomics Program, NIH grant 5U01ES017154-02 (to M.H., M.A.M., J.C. and T.T.), Cancer Research UK (to C. Caldas and P.D.P.) and the National Institute of General Medical Sciences (R01GM084875 to W.W.W.), the Canadian Breast Cancer Research Alliance and the Canadian Cancer Society (to S.A. and C.E.). We thank B. Reva, Y. Antipin and C. Sander (Memorial Sloan Kettering Cancer Center) for assistance with MutationAssessor, and G. Wu (Ontario Institute for Cancer Research) for assistance with Reactome.
Author information
Authors and Affiliations
Contributions
S.A., S.P.S., C. Caldas and M.A.M. designed and implemented the research plan and wrote the manuscript. S.P.S., A.R., R. Goya, G. Ha, J.D., G. Haffari, A. Bashashati, A. McPherson, K.S., A.C., R. Giuliany, A.H.-M., J.R., D.L., I.B., R.V., S.W.C., M.G., I.M.M., S.J., C. Curtis, O.M.R., P.D.P., V.B. and W.W.W. conducted bioinformatic analyses of the data and/or gave advice on analytic methodology. G.T. conducted histopathological review and immunohistohemistry. A.O., Y.Z., G.T., K.T., L.M.P., J.K., A.B., D.Y., A.T., N.D., T.Z., S.-F.C., K.M. and M.H. conducted sequencing or experimental validation of somatic aberrations. D.Y., A. Moradian, S.-W.G.C. and G.B.M. conducted proteome validation of splicing. P.W., K.G., S.C., S.-F.C., G.T., J.M., C. Caldas, P.D.P. and D.H. collected and interpreted clinical data. S.D., J.F.C., T.T., M.S., P.G. and C.J.E. contributed materials or reagents. K.H., V.T., T.H., M.H. and M.A.M. generated sequence data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains legends for Supplementary Figures 1-12 (see pages 2-5), Supplementary Figures 1-12 (see pages 6-127), Supplementary Methods with additional references (see pages 128-140) and legends for Supplementary Tables 1-17 with additional references (see pages 142-145). (PDF 22432 kb)
Supplementary Data
This file contains Supplementary Tables 1-17. (ZIP 5091 kb)
Rights and permissions
About this article
Cite this article
Shah, S., Roth, A., Goya, R. et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486, 395–399 (2012). https://doi.org/10.1038/nature10933
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10933
This article is cited by
-
Mutational landscape of inflammatory breast cancer
Journal of Translational Medicine (2024)
-
Prognosis prediction and risk stratification of breast cancer patients based on a mitochondria-related gene signature
Scientific Reports (2024)
-
Circulating tumour mutation detection in triple-negative breast cancer as an adjunct to tissue response assessment
npj Breast Cancer (2024)
-
Eribulin promotes proliferation of CD8+ T cells and potentiates T cell-mediated anti-tumor activity against triple-negative breast cancer cells
Breast Cancer Research and Treatment (2024)
-
Syndecan-1 as an immunogene in Triple-negative breast cancer: regulation tumor-infiltrating lymphocyte in the tumor microenviroment and EMT by TGFb1/Smad pathway
Cancer Cell International (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.