Abstract
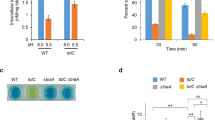
Food-borne hemorrhagic Escherichia coli, exemplified by the strains O157:H7 and O104:H4 (refs 1, 2), require elaborate acid-resistance systems (ARs)3 to survive the extremely acidic environment such as the stomach (pH ≈ 2). AR2 expels intracellular protons through the decarboxylation of l-glutamate (Glu) in the cytoplasm and exchange of the reaction product γ-aminobutyric acid (GABA) with extracellular Glu. The latter process is mediated by the Glu–GABA antiporter GadC4,5, a representative member of the amino-acid–polyamine–organocation superfamily of membrane transporters. The functional mechanism of GadC remains largely unknown. Here we show, with the use of an in vitro proteoliposome-based assay, that GadC transports GABA/Glu only under acidic conditions, with no detectable activity at pH values higher than 6.5. We determined the crystal structure of E. coli GadC at 3.1 Å resolution under basic conditions. GadC, comprising 12 transmembrane segments (TMs), exists in a closed state, with its carboxy-terminal domain serving as a plug to block an otherwise inward-open conformation. Structural and biochemical analyses reveal the essential transport residues, identify the transport path and suggest a conserved transport mechanism involving the rigid-body rotation of a helical bundle for GadC and other amino acid antiporters.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Riley, L. W. et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308, 681–685 (1983)
Rohde, H. et al. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N. Engl. J. Med. 365, 718–724 (2011)
Foster, J. W. Escherichia coli acid resistance: tales of an amateur acidophile. Nature Rev. Microbiol. 2, 898–907 (2004)
Hersh, B. M., Farooq, F. T., Barstad, D. N., Blankenhorn, D. L. & Slonczewski, J. L. A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 178, 3978–3981 (1996)
Castanie-Cornet, M. P., Penfound, T. A., Smith, D., Elliott, J. F. & Foster, J. W. Control of acid resistance in Escherichia coli. J. Bacteriol. 181, 3525–3535 (1999)
Iyer, R., Williams, C. & Miller, C. Arginine–agmatine antiporter in extreme acid resistance in Escherichia coli. J. Bacteriol. 185, 6556–6561 (2003)
Gong, S., Richard, H. & Foster, J. W. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J. Bacteriol. 185, 4402–4409 (2003)
Gao, X. et al. Structure and mechanism of an amino acid antiporter. Science 324, 1565–1568 (2009)
Gao, X. et al. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature 463, 828–832 (2010)
Fang, Y. et al. Structure of a prokaryotic virtual proton pump at 3.2 Å resolution. Nature 460, 1040–1043 (2009)
Kowalczyk, L. et al. Molecular basis of substrate-induced permeation by an amino acid antiporter. Proc. Natl Acad. Sci. USA 108, 3935–3940 (2011)
Yamashita, A., Singh, S. K., Kawate, T., Jin, Y. & Gouaux, E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215–223 (2005)
Ressl, S., Terwisscha van Scheltinga, A. C., Vonrhein, C., Ott, V. & Ziegler, C. Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature 458, 47–52 (2009)
Faham, S. et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321, 810–814 (2008)
Weyand, S. et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science 322, 709–713 (2008)
Shaffer, P. L., Goehring, A., Shankaranarayanan, A. & Gouaux, E. Structure and mechanism of a Na+-independent amino acid transporter. Science 325, 1010–1014 (2009)
Fang, Y., Kolmakova-Partensky, L. & Miller, C. A bacterial arginine–agmatine exchange transporter involved in extreme acid resistance. J. Biol. Chem. 282, 176–182 (2007)
Sasahara, A., Nanatani, K., Enomoto, M., Kuwahara, S. & Abe, K. Substrate specificity of the aspartate:alanine antiporter (AspT) of Tetragenococcus halophilus in reconstituted liposomes. J. Biol. Chem. 286, 29044–29052 (2011)
Robertson, J. L., Kolmakova-Partensky, L. & Miller, C. Design, function and structure of a monomeric CIC transporter. Nature 468, 844–847 (2010)
Waterman, S. R. & Small, P. L. Identification of sigma S-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol. Microbiol. 21, 925–940 (1996)
Kobashi, K. Catalytic oxidation of sulfhydryl groups by o-phenanthroline copper complex. Biochim. Biophys. Acta 158, 239–245 (1968)
Abramson, J. et al. The lactose permease of Escherichia coli: overall structure, the sugar-binding site and the alternating access model for transport. FEBS Lett. 555, 96–101 (2003)
Jardetzky, O. Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966)
Shimamura, T. et al. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science 328, 470–473 (2010)
Watanabe, A. et al. The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature 468, 988–991 (2010)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002)
DeLano, W. L. The PyMOL Molecular Graphics System, 〈http://www.pymol.org〉 (2002)
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006)
Bailey, S. The Ccp4 suite—programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994)
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Cowtan, K. dm: an automated procedure for phase improvement by density modification. CCP4 ESF-EACBM Newsl. Protein Crystallogr. 31, 34–38 (1994)
Veenhoff, L. M. & Poolman, B. Substrate recognition at the cytoplasmic and extracellular binding site of the lactose transport protein of Streptococcus thermophilus. J. Biol. Chem. 274, 33244–33250 (1999)
Acknowledgements
We thank S. Huang and J. He (at SSRF beamline BL17U) and N. Shimizu, T. Kumasaka and S. Baba at the Spring-8 beamline BL41XU for on-site assistance, and N. Yan for discussion and comments on the manuscript. This work was supported by funds from the Ministry of Science and Technology (grant 2009CB918801), National Natural Science Foundation, and Beijing Municipal Commissions of Education and Science and Technology.
Author information
Authors and Affiliations
Contributions
D.M., P.L. and Y.S. designed all experiments. D.M., P.L., C.Y., C.F. and P.Y. performed the experiments. D.M., P.L., C.Y., C.F., P.Y., J.W. and Y.S. analysed the data. D.M., P.L., C.Y., J.W. and Y.S. contributed to manuscript preparation. Y.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains supplementary Tables 1-2, Supplementary Figures 1-14, a Supplementary Discussion and additional references. (PDF 1727 kb)
Rights and permissions
About this article
Cite this article
Ma, D., Lu, P., Yan, C. et al. Structure and mechanism of a glutamate–GABA antiporter. Nature 483, 632–636 (2012). https://doi.org/10.1038/nature10917
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10917
This article is cited by
-
Cryo-EM structure of the human Asc-1 transporter complex
Nature Communications (2024)
-
Acid-tolerant bacteria and prospects in industrial and environmental applications
Applied Microbiology and Biotechnology (2023)
-
Division of labor and collective functionality in Escherichia coli under acid stress
Communications Biology (2022)
-
Transcriptomics reveal different metabolic strategies for acid resistance and gamma-aminobutyric acid (GABA) production in select Levilactobacillus brevis strains
Microbial Cell Factories (2021)
-
Reconstruction of the glutamate decarboxylase system in Lactococcus lactis for biosynthesis of food-grade γ-aminobutyric acid
Applied Microbiology and Biotechnology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.