Abstract
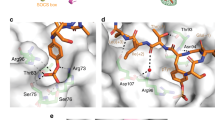
Ubiquitin (Ub) and ubiquitin-like (Ubl) modifiers such as SUMO (also known as Smt3 in Saccharomyces cerevisiae) mediate signal transduction through post-translational modification of substrate proteins in pathways that control differentiation, apoptosis and the cell cycle, and responses to stress such as the DNA damage response. In yeast, the proliferating cell nuclear antigen PCNA (also known as Pol30) is modified by ubiquitin in response to DNA damage and by SUMO during S phase. Whereas Ub–PCNA can signal for recruitment of translesion DNA polymerases, SUMO–PCNA signals for recruitment of the anti-recombinogenic DNA helicase Srs2. It remains unclear how receptors such as Srs2 specifically recognize substrates after conjugation to Ub and Ubls. Here we show, through structural, biochemical and functional studies, that the Srs2 carboxy-terminal domain harbours tandem receptor motifs that interact independently with PCNA and SUMO and that both motifs are required to recognize SUMO–PCNA specifically. The mechanism presented is pertinent to understanding how other receptors specifically recognize Ub- and Ubl-modified substrates to facilitate signal transduction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kirkin, V. & Dikic, I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr. Opin. Cell Biol. 19, 199–205 (2007)
Gareau, J. R. & Lima, C. D. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nature Rev. Mol. Cell Biol. 11, 861–871 (2010)
Dikic, I., Wakatsuki, S. & Walters, K. J. Ubiquitin-binding domains – from structures to functions. Nature Rev. Mol. Cell Biol. 10, 659–671 (2009)
Moldovan, G. L., Pfander, B. & Jentsch, S. PCNA, the maestro of the replication fork. Cell 129, 665–679 (2007)
Krishna, T. S., Kong, X. P., Gary, S., Burgers, P. M. & Kuriyan, J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79, 1233–1243 (1994)
Gulbis, J. M., Kelman, Z., Hurwitz, J., O’Donnell, M. & Kuriyan, J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87, 297–306 (1996)
Bruning, J. B. & Shamoo, Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-δ p66 subunit and flap endonuclease-1. Structure 12, 2209–2219 (2004)
Vijayakumar, S. et al. The C-terminal domain of yeast PCNA is required for physical and functional interactions with Cdc9 DNA ligase. Nucleic Acids Res. 35, 1624–1637 (2007)
Scott, M. T., Morrice, N. & Ball, K. L. Reversible phosphorylation at the C-terminal regulatory domain of p21(Waf1/Cip1) modulates proliferating cell nuclear antigen binding. J. Biol. Chem. 275, 11529–11537 (2000)
Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G. & Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 (2002)
Stelter, P. & Ulrich, H. D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425, 188–191 (2003)
Pfander, B., Moldovan, G. L., Sacher, M., Hoege, C. & Jentsch, S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 (2005)
Papouli, E. et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19, 123–133 (2005)
Krejci, L. et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 (2003)
Veaute, X. et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–312 (2003)
Lawrence, C. W. & Christensen, R. B. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol. 139, 866–876 (1979)
Baba, D. et al. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 435, 979–982 (2005)
Ulrich, H. D. PCNASUMO and Srs2: a model SUMO substrate-effector pair. Biochem. Soc. Trans. 35, 1385–1388 (2007)
Song, J., Durrin, L. K., Wilkinson, T. A., Krontiris, T. G. & Chen, Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl Acad. Sci. USA 101, 14373–14378 (2004)
Chang, C. C. et al. Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol. Cell 42, 62–74 (2011)
Hishiki, A. et al. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J. Biol. Chem. 284, 10552–10560 (2009)
Yunus, A. A. & Lima, C. D. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 35, 669–682 (2009)
Yunus, A. A. & Lima, C. D. Purification of SUMO conjugating enzymes and kinetic analysis of substrate conjugation. Methods Mol. Biol. 497, 167–186 (2009)
Freudenthal, B. D., Brogie, J. E., Gakhar, L., Kondratick, C. M. & Washington, M. T. Crystal structure of SUMO-modified proliferating cell nuclear antigen. J. Mol. Biol. 406, 9–17 (2011)
Kazmirski, S. L., Zhao, Y., Bowman, G. D., O’Donnell, M. & Kuriyan, J. Out-of-plane motions in open sliding clamps: molecular dynamics simulations of eukaryotic and archaeal proliferating cell nuclear antigen. Proc. Natl Acad. Sci. USA 102, 13801–13806 (2005)
Miyata, T. et al. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc. Natl Acad. Sci. USA 102, 13795–13800 (2005)
Kelch, B. A., Makino, D. L., O’Donnell, M. & Kuriyan, J. How a DNA polymerase clamp loader opens a sliding clamp. Science 334, 1675–1680 (2011)
Bowman, G. D., O’Donnell, M. & Kuriyan, J. Structural analysis of a eukaryotic sliding DNA clamp–clamp loader complex. Nature 429, 724–730 (2004)
Sakurai, S. et al. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 24, 683–693 (2005)
Reverter, D. & Lima, C. D. Insights into E3 ligase activity revealed by a SUMO–RanGAP1–Ubc9–Nup358 complex. Nature 435, 687–692 (2005)
Baba, D. et al. Crystal structure of SUMO-3-modified thymine-DNA glycosylase. J. Mol. Biol. 359, 137–147 (2006)
Olsen, S. K., Capili, A. D., Lu, X., Tan, D. S. & Lima, C. D. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature 463, 906–912 (2010)
Song, J., Zhang, Z., Hu, W. & Chen, Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 280, 40122–40129 (2005)
Sekiyama, N. et al. Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J. Biol. Chem. 283, 35966–35975 (2008)
Moldovan, G. L., Pfander, B. & Jentsch, S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell 23, 723–732 (2006)
Seet, B. T., Dikic, I., Zhou, M. M. & Pawson, T. Reading protein modifications with interaction domains. Nature Rev. Mol. Cell Biol. 7, 473–483 (2006)
Bienko, M. et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310, 1821–1824 (2005)
Chen, J., Ai, Y., Wang, J., Haracska, L. & Zhuang, Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nature Chem. Biol. 6, 270–272 (2010)
Moldovan, G. L. et al. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell 45, 75–86 (2012)
Mossessova, E. & Lima, C. D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 (2000)
Rayment, I. Reductive alkylation of lysine residues to alter crystallization properties of proteins. Methods Enzymol. 276, 171–179 (1997)
Otwinowski, Z. & Minor, W. in Methods in Enzymology vol. 276 (eds Carter, C. W. Jr. & Sweet, R. M. ) 307–326 (Academic Press, 1997)
Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Vagin, A. & Teplyakov, A. MOLREP: an automated program from molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Delano, W. The PyMOL Molecular Graphics System (DeLano Scientific, 2002)
Acknowledgements
We thank J. Kuriyan and B. Kelch for coordinates of the T4 clamp before publication. NE-CAT beamlines (Advanced Photon Source) supported by RR-15301 (NIH NCRR). APS supported by the US Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Beamline X29 (National Synchrotron Light Source) supported by the US Department of Energy, the Office of Basic Energy Sciences and P41RR012408 (NIH NCRR). A.A.A., F.M. and C.D.L. are supported by NIH R01 GM065872 to C.D.L. and F32 GM086066 to A.A.A.
Author information
Authors and Affiliations
Contributions
Experiments performed and analysed by A.A.A., F.M. and C.D.L. Manuscript prepared by A.A.A and C.D.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-2 and Supplementary Figures 1-14 with legends. (PDF 1709 kb)
Rights and permissions
About this article
Cite this article
Armstrong, A., Mohideen, F. & Lima, C. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 483, 59–63 (2012). https://doi.org/10.1038/nature10883
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10883
This article is cited by
-
An expanded lexicon for the ubiquitin code
Nature Reviews Molecular Cell Biology (2023)
-
Activity-based profiling of cullin–RING E3 networks by conformation-specific probes
Nature Chemical Biology (2023)
-
A Critical Role for ISGylation, Ubiquitination and, SUMOylation in Brain Damage: Implications for Neuroprotection
Neurochemical Research (2020)
-
The PCNA interaction motifs revisited: thinking outside the PIP-box
Cellular and Molecular Life Sciences (2019)
-
Crystal structure and SUMO binding of Slx1-Slx4 complex
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.