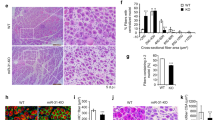
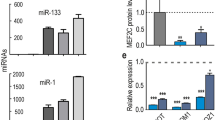
Abstract
Among the key properties that distinguish adult mammalian stem cells from their more differentiated progeny is the ability of stem cells to remain in a quiescent state for prolonged periods of time1,2. However, the molecular pathways for the maintenance of stem-cell quiescence remain elusive. Here we use adult mouse muscle stem cells (satellite cells) as a model system and show that the microRNA (miRNA) pathway is essential for the maintenance of the quiescent state. Satellite cells that lack a functional miRNA pathway spontaneously exit quiescence and enter the cell cycle. We identified quiescence-specific miRNAs in the satellite-cell lineage by microarray analysis. Among these, miRNA-489 (miR-489) is highly expressed in quiescent satellite cells and is quickly downregulated during satellite-cell activation. Further analysis revealed that miR-489 functions as a regulator of satellite-cell quiescence, as it post-transcriptionally suppresses the oncogene Dek, the protein product of which localizes to the more differentiated daughter cell during asymmetric division of satellite cells and promotes the transient proliferative expansion of myogenic progenitors. Our results provide evidence of the miRNA pathway in general, and of a specific miRNA, miR-489, in actively maintaining the quiescent state of an adult stem-cell population.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Li, L. & Clevers, H. Coexistence of quiescent and active adult stem cells in mammals. Science 327, 542–545 (2010)
Fuchs, E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell 137, 811–819 (2009)
Yi, R., Poy, M. N., Stoffel, M. & Fuchs, E. A skin microRNA promotes differentiation by repressing 'stemness'. Nature 452, 225–229 (2008)
Tiscornia, G. & Izpisua Belmonte, J. C. MicroRNAs in embryonic stem cell function and fate. Genes Dev. 24, 2732–2741 (2010)
Nishijo, K. et al. Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J. 23, 2681–2690 (2009)
Harfe, B. D., McManus, M. T., Mansfield, J. H., Hornstein, E. & Tabin, C. J. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl Acad. Sci. USA 102, 10898–10903 (2005)
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).f
Morgan, J. E. & Partridge, T. A. Muscle satellite cells. Int. J. Biochem. Cell Biol. 35, 1151–1156 (2003)
Friedman, R. C., Farh, K. K., Burge, C. B. & Bartel, D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 (2009)
Fukada, S. et al. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells 25, 2448–2459 (2007)
van Rooij, E. et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579 (2007)
Olguin, H. C. & Olwin, B. B. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev. Biol. 275, 375–388 (2004)
Tanaka, K. K. et al. Syndecan-4-expressing muscle progenitor cells in the SP engraft as satellite cells during muscle regeneration. Cell Stem Cell 4, 217–225 (2009)
Zammit, P. S., Partridge, T. A. & Yablonka-Reuveni, Z. The skeletal muscle satellite cell: the stem cell that came in from the cold. J. Histochem. Cytochem. 54, 1177–1191 (2006)
Krutzfeldt, J. et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature 438, 685–689 (2005)
Grimson, A. et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 (2007)
Khodadoust, M. S. et al. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 69, 6405–6413 (2009)
Soares, L. M., Zanier, K., Mackereth, C., Sattler, M. & Valcarcel, J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science 312, 1961–1965 (2006)
Zammit, P. S. et al. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J. Cell Biol. 166, 347–357 (2004)
Zammit, P. S. et al. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 119, 1824–1832 (2006)
Conboy, M. J., Karasov, A. O. & Rando, T. A. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 5, e102 (2007)
Shinin, V., Gayraud-Morel, B., Gomes, D. & Tajbakhsh, S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nature Cell Biol. 8, 677–687 (2006)
Rosenblatt, J. D., Lunt, A. I., Parry, D. J. & Partridge, T. A. Culturing satellite cells from living single muscle fibre explants. In vitro Cell Dev. Biol. Anim. 31, 773–779 (1995)
Bertoni, C. et al. Enhancement of plasmid-mediated gene therapy for muscular dystrophy by directed plasmid integration. Proc. Natl Acad. Sci. USA 103, 419–424 (2006)
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45 (2001)
Acknowledgements
We thank the members of the Rando laboratory for comments and discussions. We thank B. Olwin for providing the syndecan 4 antibody. This work was supported by the Glenn Foundation for Medical Research and by grants from the National Institutes of Health (NIH) (P01 AG036695, R01 AG23806 (R37 MERIT Award), R01 AR062185 and DP1 OD000392 (an NIH Director's Pioneer Award)) and the Department of Veterans Affairs (Merit Review) to T.A.R.
Author information
Authors and Affiliations
Contributions
T.H.C. and T.A.R. conceived the study. T.H.C., N.L.Q., G.W.C., L.L. and T.A.R. designed the experiments. T.H.C., B.Y. and L.L. performed all FACS analyses. T.H.C., N.L.Q., G.W.C., L.P., A.E., B.Y. and P.H. performed the experiments and analysed the experimental data. T.H.C. and T.A.R. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-14 with legends and Supplementary Table 1. (PDF 2023 kb)
Rights and permissions
About this article
Cite this article
Cheung, T., Quach, N., Charville, G. et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 482, 524–528 (2012). https://doi.org/10.1038/nature10834
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10834
This article is cited by
-
Regulation of muscle stem cell fate
Cell Regeneration (2022)
-
CPEB1 directs muscle stem cell activation by reprogramming the translational landscape
Nature Communications (2022)
-
Control of satellite cell function in muscle regeneration and its disruption in ageing
Nature Reviews Molecular Cell Biology (2022)
-
Key concepts in muscle regeneration: muscle “cellular ecology” integrates a gestalt of cellular cross-talk, motility, and activity to remodel structure and restore function
European Journal of Applied Physiology (2022)
-
Muscle Stem Cell Function Is Impaired in β2-Adrenoceptor Knockout Mice
Stem Cell Reviews and Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.