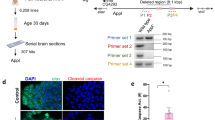
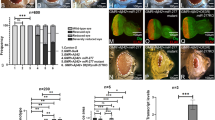
Abstract
Human neurodegenerative diseases have the temporal hallmark of afflicting the elderly population. Ageing is one of the most prominent factors to influence disease onset and progression1, yet little is known about the molecular pathways that connect these processes. To understand this connection it is necessary to identify the pathways that functionally integrate ageing, chronic maintenance of the brain and modulation of neurodegenerative disease. MicroRNAs (miRNA) are emerging as critical factors in gene regulation during development; however, their role in adult-onset, age-associated processes is only beginning to be revealed. Here we report that the conserved miRNA miR-34 regulates age-associated events and long-term brain integrity in Drosophila, providing a molecular link between ageing and neurodegeneration. Fly mir-34 expression exhibits adult-onset, brain-enriched and age-modulated characteristics. Whereas mir-34 loss triggers a gene profile of accelerated brain ageing, late-onset brain degeneration and a catastrophic decline in survival, mir-34 upregulation extends median lifespan and mitigates neurodegeneration induced by human pathogenic polyglutamine disease protein. Some of the age-associated effects of miR-34 require adult-onset translational repression of Eip74EF, an essential ETS domain transcription factor involved in steroid hormone pathways. Our studies indicate that miRNA-dependent pathways may have an impact on adult-onset, age-associated events by silencing developmental genes that later have a deleterious influence on adult life cycle and disease, and highlight fly miR-34 as a key miRNA with a role in this process.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Amaducci, L. & Tesco, G. Aging as a major risk for degenerative diseases of the central nervous system. Curr. Opin. Neurol. 7, 283–286 (1994)
Eacker, S. M., Dawson, T. M. & Dawson, V. L. Understanding microRNAs in neurodegeneration. Nature Rev. Neurosci. 10, 837–841 (2009)
Bilen, J., Liu, N., Burnett, B. G., Pittman, R. N. & Bonini, N. M. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol. Cell 24, 157–163 (2006)
Jiang, F. et al. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 19, 1674–1679 (2005)
Liu, N. et al. The exoribonuclease Nibbler controls 3′ end processing of microRNAs in Drosophila. Curr. Biol. 21, 1888–1893 (2011)
Han, B. W. H. u. n. g. J. H., Weng, Z., Zamore, P. D. & Ameres, S. L. The 3′-to-5′ exoribonuclease Nibbler shapes the 3′ ends of microRNAs bound to Drosophila Argonaute1. Curr. Biol. 21, 1878–1887 (2011)
Chung, W. J., Okamura, K., Martin, R. & Lai, E. C. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 18, 795–802 (2008)
Kretzschmar, D., Hasan, G., Sharma, S., Heisenberg, M. & Benzer, S. The swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. J. Neurosci. 17, 7425–7432 (1997)
Cao, K., Chen-Plotkin, A. S., Plotkin, J. B. & Wang, L. S. Age-correlated gene expression in normal and neurodegenerative human brain tissues. PLoS ONE 5, (2010)
Simon, A. F., Shih, C., Mack, A. & Benzer, S. Steroid control of longevity in Drosophila melanogaster. Science 299, 1407–1410 (2003)
Fletcher, J. C. & Thummel, C. S. The Drosophila E74 gene is required for the proper stage- and tissue-specific transcription of ecdysone-regulated genes at the onset of metamorphosis. Development 121, 1411–1421 (1995)
Fletcher, J. C., D’Avino, P. P. & Thummel, C. S. A steroid-triggered switch in E74 transcription factor isoforms regulates the timing of secondary-response gene expression. Proc. Natl Acad. Sci. USA 94, 4582–4586 (1997)
Morimoto, R. I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22, 1427–1438 (2008)
Warrick, J. M. et al. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell 93, 939–949 (1998)
Hirth, F. Drosophila melanogaster in the study of human neurodegeneration. CNS Neurol. Disord. Drug Targets 9, 504–523 (2010)
Williams, G. C. Pleitropy, natural selection and the evolution of senescence. Evolution 11, 398–411 (1957)
Kirkwood, T. B. Understanding the odd science of aging. Cell 120, 437–447 (2005)
Boehm, M. & Slack, F. A developmental timing microRNA and its target regulate life span in C. elegans. Science 310, 1954–1957 (2005)
de Lencastre, A. et al. MicroRNAs both promote and antagonize longevity in C. elegans. Curr. Biol. 20, 2159–2168 (2010)
Ibanez-Ventoso, C. et al. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell 5, 235–246 (2006)
Bak, M. et al. MicroRNA expression in the adult mouse central nervous system. RNA 14, 432–444 (2008)
Zovoilis, A. et al. microRNA-34c is a novel target to treat dementias. EMBO J. 30, 4299–4308 (2011)
Minones-Moyano, E. et al. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 20, 3067–3078 (2011)
Li, X., Khanna, A., Li, N. & Wang, E. Circulatory miR34a as an RNA based, noninvasive biomarker for brain aging. Aging 3, 985–1002 (2011)
Khanna, A., Muthusamy, S., Liang, R., Sarojini, H. & Wang, E. Gain of survival signaling by down-regulation of three key miRNAs in brain of calorie-restricted mice. Aging 3, 223–236 (2011)
Gaughwin, P. M. et al. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington’s disease. Hum. Mol. Genet. 20, 2225–2237 (2011)
Yang, J. et al. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age 10.1007/s11357-011-9324-3 (2011)
Parks, A. L. et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nature Genet. 36, 288–292 (2004)
Li, L. B., Yu, Z., Teng, X. & Bonini, N. M. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature 453, 1107–1111 (2008)
Dockendorff, T. C. et al. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron 34, 973–984 (2002)
Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P. & Burge, C. B. Prediction of mammalian microRNA targets. Cell 115, 787–798 (2003)
Grün, D., Wang, Y. L., Langenberger, D., Gunsalus, K. C. & Rajewsky, N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLOS Comput. Biol. 1, e13 (2005)
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004)
Acknowledgements
We thank C. Thummel, T. Jongens and A. Bashirullah for reagents. We are grateful to A. Cashmore, A. Burguete, J. Kim, S. Cherry, B. Gregory, A. Gitler and the Bonini laboratory for discussion and critical reading of the manuscript. We thank X. Teng for assistance with fly paraffin section. This work was funded by the NINDS (R01-NS043578) and the Ellison Foundation (to N.M.B.). L.-S.W. and K.C. are supported by a pilot grant from Penn Genome Frontiers Institute. L.-S.W. is supported by NIA (U01-AG-032984-02 and RC2-AG036528-01) and a Penn Institute on Aging pilot grant (AG010124). N.M.B. is an Investigator of the Howard Hughes Medical Institute. J.R.K. received support from NIH T32 AG00255.
Author information
Authors and Affiliations
Contributions
N.L. and N.M.B. conceived and designed the project. N.L., M.L., M.A., G.-J.H., J.R.K. and Y.Z. planned, executed and analysed experiments. K.C. and L.S.-W. performed aging computational modelling. N.L. and N.M.B. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-8 with legends, Supplementary Table 1 and additional references. (PDF 5144 kb)
Supplementary Table 2
This table contains the age-correlated probesets. The dd*dir value describes if a particular gene is upregulated (positive value) or downregulated (negative value) as well as the slope of the change compared to the diagonal. In miR-34 mutants, a large number of probesets have a positive dd*dir value, meaning they show higher expression and change faster compared to age-matched controls. (XLS 74 kb)
Supplementary Table 3
This table contains the DAVID functional analysis of probesets positively and negatively correlated with age. DAVID functional analysis of the probesets positively and negatively correlated with age (See Supplementary Table S2) extracted from microarray analysis of control animal brains. These two sets were enriched for different functional terms. Terms with significance p<0.001 are listed. (XLS 28 kb)
Supplementary Table 4
This table contains a summary of lifespan results. For lifespan analysis, flies were generated in the same uniform homogeneous genetic background, 5905. Log-rank test was used for statistics analysis. (XLS 24 kb)
Rights and permissions
About this article
Cite this article
Liu, N., Landreh, M., Cao, K. et al. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 482, 519–523 (2012). https://doi.org/10.1038/nature10810
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10810
This article is cited by
-
Regulation of insect behavior by non-coding RNAs
Science China Life Sciences (2024)
-
miR-34a/DRP-1-mediated mitophagy participated in cisplatin-induced ototoxicity via increasing oxidative stress
BMC Pharmacology and Toxicology (2023)
-
Eip74EF is a dominant modifier for ALS-FTD-linked VCPR152H phenotypes in the Drosophila eye model
BMC Research Notes (2023)
-
The role of microRNAs in neurodegenerative diseases: a review
Cell Biology and Toxicology (2023)
-
Analysis of Aedes aegypti microRNAs in response to Wolbachia wAlbB infection and their potential role in mosquito longevity
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.