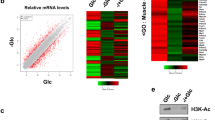
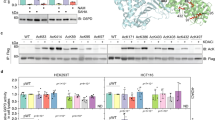
Abstract
First identified as histone-modifying proteins, lysine acetyltransferases (KATs) and deacetylases (KDACs) antagonize each other through modification of the side chains of lysine residues in histone proteins1. Acetylation of many non-histone proteins involved in chromatin, metabolism or cytoskeleton regulation were further identified in eukaryotic organisms2,3,4,5,6, but the corresponding enzymes and substrate-specific functions of the modifications are unclear. Moreover, mechanisms underlying functional specificity of individual KDACs7 remain enigmatic, and the substrate spectra of each KDAC lack comprehensive definition. Here we dissect the functional specificity of 12 critical human KDACs using a genome-wide synthetic lethality screen8,9,10,11,12,13 in cultured human cells. The genetic interaction profiles revealed enzyme–substrate relationships between individual KDACs and many important substrates governing a wide array of biological processes including metabolism, development and cell cycle progression. We further confirmed that acetylation and deacetylation of the catalytic subunit of the adenosine monophosphate-activated protein kinase (AMPK), a critical cellular energy-sensing protein kinase complex, is controlled by the opposing catalytic activities of HDAC1 and p300. Deacetylation of AMPK enhances physical interaction with the upstream kinase LKB1, leading to AMPK phosphorylation and activation, and resulting in lipid breakdown in human liver cells. These findings provide new insights into previously underappreciated metabolic regulatory roles of HDAC1 in coordinating nutrient availability and cellular responses upstream of AMPK, and demonstrate the importance of high-throughput genetic interaction profiling to elucidate functional specificity and critical substrates of individual human KDACs potentially valuable for therapeutic applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
Microarray data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE29662.
References
Yang, X. J. & Seto, E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nature Rev. Mol. Cell Biol. 9, 206–218 (2008)
Lin, Y. Y. et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136, 1073–1084 (2009)
Choudhary, C. et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 (2009)
Kim, S. C. et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 (2006)
Zhao, S. et al. Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 (2010)
Wellen, K. E. et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009)
Haberland, M., Montgomery, R. L. & Olson, E. N. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Rev. Genet. 10, 32–42 (2009)
Scholl, C. et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 137, 821–834 (2009)
Luo, J. et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835–848 (2009)
Silva, J. M. et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science 319, 617–620 (2008)
Schlabach, M. R. et al. Cancer proliferation gene discovery through functional genomics. Science 319, 620–624 (2008)
Luo, B. et al. Highly parallel identification of essential genes in cancer cells. Proc. Natl Acad. Sci. USA 105, 20380–20385 (2008)
Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009)
Stark, C. et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 39, D698–D704 (2011)
Haberland, M., Johnson, A., Mokalled, M. H., Montgomery, R. L. & Olson, E. N. Genetic dissection of histone deacetylase requirement in tumor cells. Proc. Natl Acad. Sci. USA 106, 7751–7755 (2009)
Lin, Y. Y. et al. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 22, 2062–2074 (2008)
Costanzo, M. et al. The genetic landscape of a cell. Science 327, 425–431 (2010)
Hassig, C. A. et al. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl Acad. Sci. USA 95, 3519–3524 (1998)
Grozinger, C. M. & Schreiber, S. L. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem. Biol. 9, 3–16 (2002)
Hardie, D. G. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nature Rev. Mol. Cell Biol. 8, 774–785 (2007)
Xiao, B. et al. Structure of mammalian AMPK and its regulation by ADP. Nature 472, 230–233 (2011)
Oakhill, J. S. et al. AMPK is a direct adenylate charge-regulated protein kinase. Science 332, 1433–1435 (2011)
Hawley, S. A. et al. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J. Biol. Chem. 271, 27879–27887 (1996)
Hawley, S. A. et al. Complexes between the LKB1 tumor suppressor, STRADα/β and MO25α/β are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2, 28 (2003)
Suter, M. et al. Dissecting the role of 5′-AMP for allosteric stimulation, activation, and deactivation of AMP-activated protein kinase. J. Biol. Chem. 281, 32207–32216 (2006)
Bungard, D. et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science 329, 1201–1205 (2010)
Bolden, J. E., Peart, M. J. & Johnstone, R. W. Anticancer activities of histone deacetylase inhibitors. Nature Rev. Drug Discov. 5, 769–784 (2006)
Kazantsev, A. G. & Thompson, L. M. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nature Rev. Drug Discov. 7, 854–868 (2008)
Huangfu, D. et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nature Biotechnol. 26, 795–797 (2008)
Bantscheff, M. et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nature Biotechnol. 29, 255–265 (2011)
Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA 95, 14863–14868 (1998)
Thomas, P. D. et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003)
Mi, H. et al. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38, D204–D210 (2010)
Berriz, G. F., Beaver, J. E., Cenik, C., Tasan, M. & Roth, F. P. Next generation software for functional trend analysis. Bioinformatics 25, 3043–3044 (2009)
Jayapandian, M. et al. Michigan Molecular Interactions (MiMI): putting the jigsaw puzzle together. Nucleic Acids Res. 35, D566–D571 (2007)
Nakatani, Y. & Ogryzko, V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370, 430–444 (2003)
Hwang, J. T. et al. Genistein, EGCG, and capsaicin inhibit adipocyte differentiation process via activating AMP-activated protein kinase. Biochem. Biophys. Res. Commun. 338, 694–699 (2005)
Saldanha, A. J. Java Treeview—extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004)
Acknowledgements
We thank H. Zhu, S.-L. Yu and L.-P. Chow for gifts of reagents, J. Lu for IMR-90 cells, Department of Medical Science, National Taiwan University for technical help with sequencing, and the National RNAi Core Facility, Academia Sinica for reagents and technical support with RNAi screen. We are grateful to P. Meluh and members of the Lin and Boeke laboratories for discussions. We thank D. Root for discussions. This study was supported by National Science Council grants NSC 98-2320-B-002-057-, NSC 99-2320-B-002-057- and NSC 100-2325-002-044- (Y.-y.L.), National Taiwan University Frontier and Innovative Research 99R71424 (Y.-y.L.), National Taiwan University College of Medicine and National Taiwan University Hospital Excellent Translational Medicine Research Project 99C101-603 (Y.-y.L. and J.-y.L.), National Health Research Institutes Career Development grant NHRI-EX100-10017BC (Y.-y.L.), Liver Disease Prevention and Treatment Research Foundation (Y.-y.L. and J.-y.L.) and NIH Common Fund grant U54 RR 020839 (J.D.B.).
Author information
Authors and Affiliations
Contributions
Y.-y.L. and J.D.B. designed the experiments, with help from J.-y.L. Y.-y.L. and Y.-h.C. performed the primary screens, with help from C.N.K. and C.-L.L. Validation and further functional categorization experiments were performed by Y.-y.L., S.-Y.L. and Z.K. S.K. and Y.S. performed computational analyses, supervised by R.I. and J.S.B., respectively. Y.-y.L., J.-y.L. and J.D.B. wrote the manuscript. All authors discussed results and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-18 with legends and Supplementary Tables 1-8. (PDF 13084 kb)
Rights and permissions
About this article
Cite this article
Lin, Yy., Kiihl, S., Suhail, Y. et al. Functional dissection of lysine deacetylases reveals that HDAC1 and p300 regulate AMPK. Nature 482, 251–255 (2012). https://doi.org/10.1038/nature10804
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10804
This article is cited by
-
Glioma-initiating cells at tumor edge gain signals from tumor core cells to promote their malignancy
Nature Communications (2020)
-
The chromatin remodeler RSF1 controls centromeric histone modifications to coordinate chromosome segregation
Nature Communications (2018)
-
Reciprocal regulation of RORγt acetylation and function by p300 and HDAC1
Scientific Reports (2015)
-
Epigenetic and Immune Regulation of Colorectal Cancer Stem Cells
Current Colorectal Cancer Reports (2015)
-
Caloric restriction mimetics: towards a molecular definition
Nature Reviews Drug Discovery (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.