Abstract
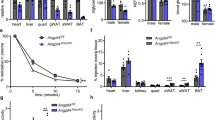
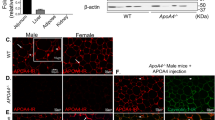
Free fatty acids provide an important energy source as nutrients, and act as signalling molecules in various cellular processes1,2,3,4. Several G-protein-coupled receptors have been identified as free-fatty-acid receptors important in physiology as well as in several diseases3,5,6,7,8,9,10,11,12,13. GPR120 (also known as O3FAR1) functions as a receptor for unsaturated long-chain free fatty acids and has a critical role in various physiological homeostasis mechanisms such as adipogenesis, regulation of appetite and food preference5,6,14,15,16. Here we show that GPR120-deficient mice fed a high-fat diet develop obesity, glucose intolerance and fatty liver with decreased adipocyte differentiation and lipogenesis and enhanced hepatic lipogenesis. Insulin resistance in such mice is associated with reduced insulin signalling and enhanced inflammation in adipose tissue. In human, we show that GPR120 expression in adipose tissue is significantly higher in obese individuals than in lean controls. GPR120 exon sequencing in obese subjects reveals a deleterious non-synonymous mutation (p.R270H) that inhibits GPR120 signalling activity. Furthermore, the p.R270H variant increases the risk of obesity in European populations. Overall, this study demonstrates that the lipid sensor GPR120 has a key role in sensing dietary fat and, therefore, in the control of energy balance in both humans and rodents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
28 February 2012
The Supplementary Tables were missing from the original supplementary information file and have since been added.
References
Nunez, E. A. Biological complexity is under the ‘strange attraction’ of non-esterified fatty acids. Prostaglandins Leukot. Essent. Fatty Acids 57, 107–110 (1997)
Haber, E. P. et al. Pleiotropic effects of fatty acids on pancreatic β-cells. J. Cell. Physiol. 194, 1–12 (2003)
Itoh, Y. et al. Free fatty acids regulate insulin secretion from pancreatic beta cells through GPR40. Nature 422, 173–176 (2003)
Cao, H. et al. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell 134, 933–944 (2008)
Hirasawa, A. et al. Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nature Med. 11, 90–94 (2005)
Steneberg, P., Rubins, N., Bartoov-Shifman, R., Walker, M. D. & Edlund, H. The FFA receptor GPR40 links hyperinsulinemia, hepatic steatosis, and impaired glucose homeostasis in mouse. Cell Metab. 1, 245–258 (2005)
Wang, J., Wu, X., Simonavicius, N., Tian, H. & Ling, L. Medium-chain fatty acids as ligands for orphan G protein-coupled receptor GPR84. J. Biol. Chem. 281, 34457–34464 (2006)
Ichimura, A., Hirasawa, A., Hara, T. & Tsujimoto, G. Free fatty acid receptors act as nutrient sensors to regulate energy homeostasis. Prostaglandins Other Lipid Mediat. 89, 82–88 (2009)
Maslowski, K. M. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 (2009)
Ahmed, K. et al. An autocrine lactate loop mediates insulin-dependent inhibition of lipolysis through GPR81. Cell Metab. 11, 311–319 (2010)
Oh. Da, Y. et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 142, 687–698 (2010)
Hara, T., Hirasawa, A., Ichimura, A., Kimura, I. & Tsujimoto, G. Free fatty acid receptors FFAR1 and GPR120 as novel therapeutic targets for metabolic disorders. J. Pharm. Sci. 100, 3594–3601 (2011)
Kimura, I. et al. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc. Natl Acad. Sci. USA 108, 8030–8035 (2011)
Gotoh, C. et al. The regulation of adipogenesis through GPR120. Biochem. Biophys. Res. Commun. 354, 591–597 (2007)
Tanaka, T. et al. Free fatty acids induce cholecystokinin secretion through GPR120. Naunyn Schmiedebergs Arch. Pharmacol. 377, 523–527 (2008)
Miyauchi, S. et al. Distribution and regulation of protein expression of the free fatty acid receptor GPR120. Naunyn Schmiedebergs Arch. Pharmacol. 379, 427–434 (2009)
Kido, Y. et al. Tissue-specific insulin resistance in mice with mutations in the insulin receptor, IRS-1, and IRS-2. J. Clin. Invest. 105, 199–205 (2000)
Bernal-Mizrachi, E., Wen, W., Stahlhut, S., Welling, C. M. & Permutt, M. A. Islet beta cell expression of constitutively active Akt1/PKB alpha induces striking hypertrophy, hyperplasia, and hyperinsulinemia. J. Clin. Invest. 108, 1631–1638 (2001)
Hosooka, T. et al. Dok1 mediates high-fat diet-induced adipocyte hypertrophy and obesity through modulation of PPAR-γ phosphorylation. Nature Med. 14, 188–193 (2008)
Ntambi, J. M. et al. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc. Natl Acad. Sci. USA 99, 11482–11486 (2002)
Gutierrez-Juarez, R. et al. Critical role of stearoyl-CoA desaturase-1 (SCD1) in the onset of diet-induced hepatic insulin resistance. J. Clin. Invest. 116, 1686–1695 (2006)
Jeyakumar, S. M. et al. Fatty acid desaturation index correlates with body mass and adiposity indices of obesity in Wistar NIN obese mutant rat strains WNIN/Ob and WNIN/GR-Ob. Nutr. Metab. (Lond.) 6, 27 (2009)
Brown, J. M. et al. Combined therapy of dietary fish oil and stearoyl-CoA desaturase 1 inhibition prevents the metabolic syndrome and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 30, 24–30 (2010)
Ichimura, A., Ruike, Y., Terasawa, K., Shimizu, K. & Tsujimoto, G. MicroRNA-34a inhibits cell proliferation by repressing mitogen-activated protein kinase kinase 1 during megakaryocytic differentiation of K562 cells. Mol. Pharmacol. 77, 1016–1024 (2010)
Hara, T. et al. Novel selective ligands for free fatty acid receptors GPR120 and GPR40. Naunyn Schmiedebergs Arch. Pharmacol. 380, 247–255 (2009)
Sun, Q. et al. Structure-activity relationships of GPR120 agonists based on a docking simulation. Mol. Pharmacol. 78, 804–810 (2010)
Gautier, L., Cope, L., Bolstad, B. M. & Irizarry, R. A. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 (2004)
Poulain-Godefroy, O., Lecoeur, C., Pattou, F., Fruhbeck, G. & Froguel, P. Inflammation is associated with a decrease of lipogenic factors in omental fat in women. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R1–R7 (2008)
Meyre, D. et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nature Genet. 41, 157–159 (2009)
Morandi, A. et al. The Q121 variant of ENPP1 may protect from childhood overweight/obesity in the Italian population. Obesity (Silver Spring) 17, 202–206 (2009)
Buzzetti, R. et al. PPAR-γ2 Pro12Ala variant is associated with greater insulin sensitivity in childhood obesity. Pediatr. Res. 57, 138–140 (2005)
Järvelin, M. R. et al. Ecological and individual predictors of birthweight in a northern Finland birth cohort 1986. Paediatr. Perinat. Epidemiol. 11, 298–312 (1997)
Peeters, A. et al. Variants in the FTO gene are associated with common obesity in the Belgian population. Mol. Genet. Metab. 93, 481–484 (2008)
Steffen, R., Potoczna, N., Bieri, N. & Horber, F. F. Successful multi-intervention treatment of severe obesity: a 7-year prospective study with 96% follow-up. Obes. Surg. 19, 3–12 (2009)
Rouskas, K. et al. Association between BBS6/MKKS gene polymorphisms, obesity and metabolic syndrome in the Greek population. Int. J. Obes. (Lond.) 32, 1618–1625 (2008)
Balkau, B. An epidemiologic survey from a network of French Health Examination Centres (D.E.S.I.R.): epidemiologic data on the insulin resistance syndrome [in French]. Rev. Epidemiol. Sante Publique 44, 373–375 (1996)
Jaquet, D., Collin, D., Levy-Marchal, C. & Czernichow, P. Adult height distribution in subjects born small for gestational age. Horm. Res. 62, 92–96 (2004)
Meyre, D. et al. R125W coding variant in TBC1D1 confers risk for familial obesity and contributes to linkage on chromosome 4p14 in the French population. Hum. Mol. Genet. 17, 1798–1802 (2008)
Jarick, I. et al. Novel common copy number variation for early onset extreme obesity on chromosome 11q11 identified by a genome-wide analysis. Hum. Mol. Genet. 20, 840–852 (2011)
Poskitt, E. M. Defining childhood obesity: the relative body mass index (BMI). European Childhood Obesity group. Acta Paediatr. 84, 961–963 (1995)
Rolland-Cachera, M. F. et al. Body mass index variations: centiles from birth to 87 years. Eur. J. Clin. Nutr. 45, 13–21 (1991)
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005)
Ramensky, V., Bork, P. & Sunyaev, S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900 (2002)
Sunyaev, S. et al. Prediction of deleterious human alleles. Hum. Mol. Genet. 10, 591–597 (2001)
Thomas, P. D. et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003)
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols 4, 1073–1081 (2009)
Bromberg, Y., Yachdav, G. & Rost, B. SNAP predicts effect of mutations on protein function. Bioinformatics 24, 2397–2398 (2008)
Ferrer-Costa, C. et al. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics 21, 3176–3178 (2005)
Acknowledgements
We are indebted to all subjects who participated in these studies. In Japan, the study was supported in part by research grants from the Japan Society for the Promotion of Science; the Ministry of Education, Culture, Sports, Science and Technology of Japan; the Japan Science and Technology Agency; and the Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program), initiated by the Council for Science and Technology Policy. A.I. is a fellow of the Japan Society for the Promotion of Science. A.B. is a fellow of the EU-funded EUROCHIP consortium. In France, the study was supported by le Conseil Régional Nord Pas de Calais/FEDER and the Agence Nationale de la Recherche (Programme de Recherche en Nutrition et Alimentation, SensoFAT). The Northern Finland Birth Cohort Studies 1986 received financial support from the Academy of Finland, the University Hospital of Oulu (Finland), the University of Oulu (Finland), the European Commission (EURO-BLCS, Framework 5 award QLG1-CT-2000-01643), and the Medical Research Council (G0500539, G0600705, PrevMetSyn/SALVE). We thank the ABOS consortium and the CIC-CCPPRB (Lille CHRU) team for their help in sample handling and clinical data collection. We are grateful to M. Deweirder and F. Allegaert for human DNA bank management.
Author information
Authors and Affiliations
Contributions
A.I., A. Hirasawa, O.P.-G. and A.B. are equally contributing first authors. G.T. and P.F. had the ideas for the mouse and human projects, respectively. A.I., A. Hirasawa, A.B., P.F. and G.T. drafted the manuscript. O.P.-G., H.C., D.M. and I.W. edited the manuscript and contributed to discussions. A. Hirasawa and G.T. designed the mouse research. A.I., A. Hirasawa, K.I. and G.T. created Gpr120-mutant mice. A.I., A. Hirasawa, A. Körner, T.H., I.K., T.-a.K., K.A., M. Takeuchi, K.O., N.L. and G.T. conducted biochemical and histochemical analyses for the mouse study. A.I. and A. Hirasawa performed bioinformatic analysis for the mouse study. L.Y. and C.L. performed the statistical analyses, and A.B. contributed to statistical analyses for the human study. O.P.-G. and I.W. designed the human expression gene study. A.L. performed the human expression gene study. H.C. and S.V. performed GPR120 sequencing and variant genotyping, respectively. P.B., M. Tauber, C.M., A.M., R.B., P.E., M.-R.J., W.V.H., L.V.G., F.H., B.B., C.L.-M., K.R., A. Kouvatsi and F.P. contributed to cohort-study samples and researched data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-7 with legends and Supplementary Tables 1-7. The Supplementary Tables were missing from the original file posted on line and were added on 28 February 2012. (PDF 2380 kb)
Rights and permissions
About this article
Cite this article
Ichimura, A., Hirasawa, A., Poulain-Godefroy, O. et al. Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nature 483, 350–354 (2012). https://doi.org/10.1038/nature10798
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10798
This article is cited by
-
GPCRs involved in metabolic diseases: pharmacotherapeutic development updates
Acta Pharmacologica Sinica (2024)
-
Characterization of free fatty acid receptor family in rainbow trout (Oncorhynchus mykiss): towards a better understanding of their involvement in fatty acid signalisation
BMC Genomics (2023)
-
The early life immune dynamics and cellular drivers at single-cell resolution in lamb forestomachs and abomasum
Journal of Animal Science and Biotechnology (2023)
-
RNA-seq analysis to identify genes related to resting egg production of panarctic Daphnia pulex
BMC Genomics (2023)
-
The Influence of the FFAR4 Agonist TUG-891 on Liver Steatosis in ApoE-Knockout Mice
Cardiovascular Drugs and Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.