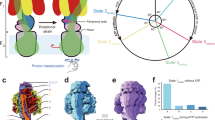
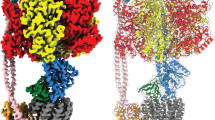
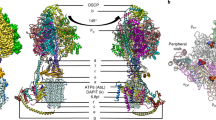
Abstract
Ion-translocating rotary ATPases serve either as ATP synthases, using energy from a transmembrane ion motive force to create the cell’s supply of ATP, or as transmembrane ion pumps that are powered by ATP hydrolysis1. The members of this family of enzymes each contain two rotary motors: one that couples ion translocation to rotation and one that couples rotation to ATP synthesis or hydrolysis. During ATP synthesis, ion translocation through the membrane-bound region of the complex causes rotation of a central rotor that drives conformational changes and ATP synthesis in the catalytic region of the complex. There are no structural models available for the intact membrane region of any ion-translocating rotary ATPase. Here we present a 9.7 Å resolution map of the H+-driven ATP synthase from Thermus thermophilus obtained by electron cryomicroscopy of single particles in ice. The 600-kilodalton complex has an overall subunit composition of A3B3CDE2FG2IL12. The membrane-bound motor consists of a ring of L subunits and the carboxy-terminal region of subunit I, which are equivalent to the c and a subunits of most other rotary ATPases, respectively. The map shows that the ring contains 12 L subunits2 and that the I subunit has eight transmembrane helices3. The L12 ring and I subunit have a surprisingly small contact area in the middle of the membrane, with helices from the I subunit making contacts with two different L subunits. The transmembrane helices of subunit I form bundles that could serve as half-channels across the membrane, with the first half-channel conducting protons from the periplasm to the L12 ring and the second half-channel conducting protons from the L12 ring to the cytoplasm. This structure therefore suggests the mechanism by which a transmembrane proton motive force is converted to rotation in rotary ATPases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
11 January 2012
The placement of an arrow was corrected in Fig. 4b.
References
Muench, S. P., Trinick, J. & Harrison, M. A. Structural divergence of the rotary ATPases. Q. Rev. Biophys. 44, 311–356 (2011)
Toei, M. et al. Dodecamer rotor ring defines H+/ATP ratio for ATP synthesis of prokaryotic V-ATPase from Thermus thermophilus. Proc. Natl Acad. Sci. USA 104, 20256–20261 (2007)
Toei, M., Toei, S. & Forgac, M. Definition of membrane topology and identification of residues important for transport in subunit a of the vacuolar ATPase. J. Biol. Chem. 286, 35176–35186 (2011)
Numoto, N., Hasegawa, Y., Takeda, K. & Miki, K. Inter-subunit interaction and quaternary rearrangement defined by the central stalk of prokaryotic V1-ATPase. EMBO Rep. 10, 1228–1234 (2009)
Lau, W. C. & Rubinstein, J. L. Structure of intact Thermus thermophilus V-ATPase by cryo-EM reveals organization of the membrane-bound V(O) motor. Proc. Natl Acad. Sci. USA 107, 1367–1372 (2010)
Abrahams, J. P., Leslie, A. G., Lutter, R. & Walker, J. E. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 (1994)
Lee, L. K. et al. The structure of the peripheral stalk of Thermus thermophilus H+-ATPase/synthase. Nature Struct. Mol. Biol. 17, 373–378 (2010)
Iwata, M. et al. Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase. Proc. Natl Acad. Sci. USA 101, 59–64 (2004)
Srinivasan, S., Vyas, N. K., Baker, M. L. & Quiocho, F. A. Crystal structure of the cytoplasmic N-terminal domain of subunit I, a homolog of subunit a, of V-ATPase. J. Mol. Biol. 412, 14–21 (2011)
Murata, T. et al. Structure of the rotor of the V-type Na+-ATPase from Enterococcus hirae. Science 308, 654–659 (2005)
Stock, D., Leslie, A. G. & Walker, J. E. Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705 (1999)
Meier, T. et al. Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science 308, 659–662 (2005)
Pogoryelov, D., Yildiz, O., Faraldo-Gomez, J. D. & Meier, T. High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nature Struct. Mol. Biol. 16, 1068–1073 (2009)
Watt, I. N. et al. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc. Natl Acad. Sci. USA 107, 16823–16827 (2010)
Preiss, L. et al. A new type of proton coordination in an F1F0-ATP synthase rotor ring. PLoS Biol. 8, e1000443 (2010)
Meier, T. et al. The central plug in the reconstituted undecameric c cylinder of a bacterial ATP synthase consists of phospholipids. FEBS Lett. 505, 353–356 (2001)
Fillingame, R. H., Angevine, C. M. & Dmitriev, O. Y. Mechanics of coupling proton movements to c-ring rotation in ATP synthase. FEBS Lett. 555, 29–34 (2003)
Junge, W., Lill, H. & Engelbrecht, S. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem. Sci. 22, 420–423 (1997)
Stouffer, A. L. et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature 451, 596–599 (2008)
Gonzales, E. B., Kawate, T. & Gouaux, E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature 460, 599–604 (2009)
Cain, B. D. & Simoni, R. D. Proton translocation by the F1F0ATPase of Escherichia coli. Mutagenic analysis of the a subunit. J. Biol. Chem. 264, 3292–3300 (1989)
Kawasaki-Nishi, S., Nishi, T. & Forgac, M. Arg-735 of the 100-kDa subunit a of the yeast V-ATPase is essential for proton translocation. Proc. Natl Acad. Sci. USA 98, 12397–12402 (2001)
Pogoryelov, D. et al. Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases. Nature Chem. Biol. 6, 891–899 (2010)
Steed, P. R. & Fillingame, R. H. Aqueous accessibility to the transmembrane regions of subunit c of the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 284, 23243–23250 (2009)
Long, J. C., Wang, S. & Vik, S. B. Membrane topology of subunit a of the F1F0 ATP synthase as determined by labeling of unique cysteine residues. J. Biol. Chem. 273, 16235–16240 (1998)
Valiyaveetil, F. I. & Fillingame, R. H. Transmembrane topography of subunit a in the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 273, 16241–16247 (1998)
Baker, L. A., Smith, E. A., Bueler, S. A. & Rubinstein, J. L. The resolution dependence of optimal exposures in liquid nitrogen temperature electron cryomicroscopy of catalase crystals. J. Struct. Biol. 169, 431–437 (2010)
Grigorieff, N. FREALIGN: high-resolution refinement of single particle structures. J. Struct. Biol. 157, 117–125 (2007)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Crowther, R. A., Henderson, R. & Smith, J. M. MRC image processing programs. J. Struct. Biol. 116, 9–16 (1996)
Nelder, J. A. & Mead, R. A simplex method for function minimization. Comput. J. 7, 308–313 (1965)
Press, W. H., Teukolsky, S. A., Vetterlin, W. T. & Flannery, B. P. Numerical Recipes in Fortran 77 2nd edn (Cambridge Univ. Press, 2003)
Sousa, D. & Grigorieff, N. Ab initio resolution measurement for single particle structures. J. Struct. Biol. 157, 201–210 (2007)
Rosenthal, P. B., Crowther, R. A. & Henderson, R. An objective criterion for resolution assessment in single-particle electron microscopy (appendix). J. Mol. Biol. 333, 743–745 (2003)
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003)
Pintilie, G. D. et al. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170, 427–438 (2010)
Baker, L. A. & Rubinstein, J. L. Edged watershed segmentation: a semi-interactive algorithm for segmentation of low-resolution maps from electron cryomicroscopy. J. Struct. Biol. 176, 127–132 (2011)
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999)
Kelley, L. A. & Sternberg, M. J. Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocols 4, 363–371 (2009)
Chacon, P. & Wriggers, W. Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol. 317, 375–384 (2002)
Goddard, T. D., Huang, C. C. & Ferrin, T. E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007)
Acknowledgements
We thank V. Kanelis, F. Sicheri, P. Rosenthal, L. Kay and R. Henderson for discussions and reading this manuscript, and J. Walker and R. Pomès for discussions. Computations were performed on the general-purpose cluster supercomputer at the SciNet HPC Consortium. W.C.Y.L. was supported by an Ontario Graduate Scholarship. J.L.R. was supported by a New Investigator Award from the Canadian Institutes of Health Research and an Early Researcher Award from the Ontario Ministry of Research and Innovation. This research was funded by operating grant MOP 81294 from the Canadian Institutes of Health Research.
Author information
Authors and Affiliations
Contributions
J.L.R. and W.C.Y.L. designed the experiments and J.L.R. supervised the research. W.C.Y.L. performed protein purification and cryo-EM. J.L.R. wrote new computer programs. W.C.Y.L. and J.L.R. performed the image analysis, interpreted the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-4 with legends, Supplementary Table 1 and additional references. (PDF 1639 kb)
Supplementary Movie 1
The movie shows a 3-D map with docked crystals structures of subunits A (yellow), B (red), C (cyan), D (blue), E (purple), F (orange), and G (beige) subunits and segments for density from subunits I (green), L (magenta), and the missing density from subunit D (blue). The scale bar corresponds to 25 Å . (MOV 20549 kb)
Rights and permissions
About this article
Cite this article
Lau, W., Rubinstein, J. Subnanometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature 481, 214–218 (2012). https://doi.org/10.1038/nature10699
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10699
This article is cited by
-
Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus
Nature Communications (2018)
-
Control of rotation of the F1FO-ATP synthase nanomotor by an inhibitory α-helix from unfolded ε or intrinsically disordered ζ and IF1 proteins
Journal of Bioenergetics and Biomembranes (2018)
-
A reciprocating motion-driven rotation mechanism for the ATP synthase
Science China Life Sciences (2016)
-
Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase
Nature (2015)
-
Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase
Nature (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.