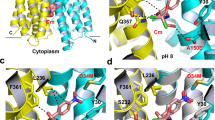
Abstract
AcrB and its homologues are the principal multidrug transporters in Gram-negative bacteria1,2,3,4,5,6 and are important in antibiotic drug tolerance7,8. AcrB is a homotrimer that acts as a tripartite complex9,10 with the outer membrane channel TolC11,12 and the membrane fusion protein AcrA13,14. Minocycline and doxorubicin have been shown to bind to the phenylalanine cluster region of the binding monomer15. Here we report the crystal structures of AcrB bound to the high-molecular-mass drugs rifampicin and erythromycin. These drugs bind to the access monomer, and the binding sites are located in the proximal multisite binding pocket, which is separated from the phenylalanine cluster region (distal pocket) by the Phe-617 loop. Our structures indicate that there are two discrete multisite binding pockets along the intramolecular channel. High-molecular-mass drugs first bind to the proximal pocket in the access state and are then forced into the distal pocket in the binding state by a peristaltic mechanism involving subdomain movements that include a shift of the Phe-617 loop. By contrast, low-molecular-mass drugs, such as minocycline and doxorubicin, travel through the proximal pocket without specific binding and immediately bind to the distal pocket. The presence of two discrete, high-volume multisite binding pockets contributes to the remarkably broad substrate recognition of AcrB.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ma, D., Cook, D. N., Hearst, J. E. & Nikaido, H. Efflux pumps and drug resistance in Gram-negative bacteria. Trends Microbiol. 2, 489–493 (1994)
Okusu, H., Ma, D. & Nikaido, H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178, 306–308 (1996)
Paulsen, I. T., Nguyen, L., Sliwinski, M. K., Rabus, R. & Saier, M. H., Jr Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301, 75–100 (2000)
Nishino, K. & Yamaguchi, A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183, 5803–5812 (2001)
Poole, K. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3, 255–264 (2001)
Nishino, K., Latifi, T. & Groisman, E. A. Virulence and drug resistance roles of multidrug efflux systems of Salmonera enterica serovar Typhimurium. Mol. Microbiol. 59, 126–141 (2006)
Poole, K., Krebes, K., McNally, C. & Neshat, S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175, 7363–7372 (1993)
Sulavik, M. C. et al. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45, 1126–1136 (2001)
Tikhonova, E. B. & Zgurskaya, H. I. AcrA, AcrB and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J. Biol. Chem. 279, 32116–32124 (2004)
Symmons, M. F., Bokma, E., Koronakis, E., Hughes, C. & Koronakis, V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl Acad. Sci. USA 106, 7173–7178 (2009)
Fralick, J. A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178, 5803–5805 (1996)
Koronakis, V., Sharff, A., Koronakis, E., Luisi, B. & Hughes, C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405, 914–919 (2000)
Zgurskaya, H. I. & Nikaido, H. AcrA is a highly asymmetric protein capable of spanning the periplasm. J. Mol. Biol. 285, 409–420 (1999)
Mikolosko, J., Bobyk, K., Zgurskaya, H. I. & Ghosh, P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure 14, 577–587 (2006)
Murakami, S., Nakashima, R., Yamashita, E., Matsumoto, T. & Yamaguchi, A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443, 173–179 (2006)
Murakami, S., Nakashima, R., Yamashita, E. & Yamaguchi, A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419, 587–593 (2002)
Yu, E. W., McDermott, G., Zgurskaya, H. I., Nikaido, H. & Koshland, D. E., Jr Structural basis of multidrug-binding capacity of the AcrB multidrug efflux pump. Science 300, 976–980 (2003)
Yu, E. W., Aires, J. R., McDermott, G. & Nikaido, H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 187, 6804–6815 (2005)
Törnroth-Horsefield, S. et al. Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist. Structure 15, 1663–1673 (2007)
Drew, D. et al. The structure of the efflux pump AcrB in complex with bile acid. Mol. Membr. Biol. 25, 677–682 (2008)
Pos, K. M., Schiefner, A., Seeger, M. A. & Diederichs, K. Crystallographic analysis of AcrB. FEBS Lett. 564, 333–339 (2004)
Murakami, S. Multidrug efflux transporter, AcrB-the pumping mechanism. Curr. Opin. Struct. Biol. 18, 459–465 (2008)
Seeger, M. A. et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313, 1295–1298 (2006)
Seeger, M. A. et al. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nature Struct. Mol. Biol. 15, 199–205 (2008)
Sennhauser, G., Amstutz, P., Briand, C., Storchenegger, O. & Grutter, M. G. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 5, e7 (2007)
Sennhauser, G., Bukowska, M. A., Briand, C. & Grutter, M. G. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J. Mol. Biol. 389, 134–145 (2009)
Schumacher, M. A. et al. Structural mechanisms of QacR induction and multidrug recognition. Science 294, 2158–2163 (2001)
Bohnert, J. A. et al. Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J. Bacteriol. 190, 8225–8229 (2008)
Medek, P., Benes, P. & Sochor, J. Computation of tunnels in protein molecules using Delaunay triangulation. J. WSCG 15, 107–114 (2007)
Pos, K. M. Drug transport mechanism of the AcrB efflux pump. Biochim. Biophys. Acta 1794, 782–793 (2009)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997)
Collaborative Computational Project. Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Brunger, A. T. Version 1.2 of the crystallography and NMR system. Nature Protocols 2, 2728–2733 (2007)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Blattner, F. R. et al. The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 (1997)
Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 (1995)
Acknowledgements
We thank T. Tsukihara for advice on the crystallographic analysis. We also thank N. Kato for discussion about the organic chemistry of the drugs that were investigated in this study. Our diffraction data were collected using Osaka University’s beamline BL44XU at SPring-8, which was equipped with an MX225-HE detector (Rayonix) and was financially supported by the Academia Sinica and the National Synchrotron Radiation Research Center (Taiwan). We are also grateful to the technical staff of the Comprehensive Analysis Center of the Institute of Scientific and Industrial Research for their assistance. This work was supported by the Program for the Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation and Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Contributions
R.N. and K.S. performed the crystallographic analysis. S.Y. and K.N. performed the molecular biological and biochemical analyses. A.Y. designed the research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-13 with legends and a Supplementary Discussion and additional references. (PDF 2173 kb)
Supplementary Movie 4a
The movie shows conformational change of the Phe-617 loop during functional rotation of AcrB. The movie was generated by morphing the three functional states of AcrB bound to both rifampicin and minocycline using PyMOL. The Phe-617 loop is shown in red. (MOV 3027 kb)
Supplementary Movie 7a
The movie shows movement of the amino acid side chains that interact with erythromycin during drug transport. The side chains of Phe 617, Ser 134 and Ser 135 are shown in red. The movie was generated by morphing the three functional states of the erythromycin-bound form of AcrB using PyMOL. (MOV 1873 kb)
Supplementary Movie 8
The movie shows crystal structure of AcrB simultaneously bound to rifampicin and minocycline solved at a 3.3 Å resolution. Rifampicin (magenta) in the access monomer (green) and minocycline (cyan) in the binding monomer (blue) are shown in a CPK (Corey-Pauling-Koltun) representation. (MOV 3426 kb)
Rights and permissions
About this article
Cite this article
Nakashima, R., Sakurai, K., Yamasaki, S. et al. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 480, 565–569 (2011). https://doi.org/10.1038/nature10641
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10641
This article is cited by
-
Functionally distinct mutations within AcrB underpin antibiotic resistance in different lifestyles
npj Antimicrobials and Resistance (2023)
-
Molecular mechanisms of antibiotic resistance revisited
Nature Reviews Microbiology (2023)
-
A role for the periplasmic adaptor protein AcrA in vetting substrate access to the RND efflux transporter AcrB
Scientific Reports (2022)
-
Allosteric drug transport mechanism of multidrug transporter AcrB
Nature Communications (2021)
-
Structure and function relationship of OqxB efflux pump from Klebsiella pneumoniae
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.