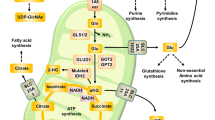
Abstract
Acetyl coenzyme A (AcCoA) is the central biosynthetic precursor for fatty-acid synthesis and protein acetylation. In the conventional view of mammalian cell metabolism, AcCoA is primarily generated from glucose-derived pyruvate through the citrate shuttle and ATP citrate lyase in the cytosol1,2,3. However, proliferating cells that exhibit aerobic glycolysis and those exposed to hypoxia convert glucose to lactate at near-stoichiometric levels, directing glucose carbon away from the tricarboxylic acid cycle and fatty-acid synthesis4. Although glutamine is consumed at levels exceeding that required for nitrogen biosynthesis5, the regulation and use of glutamine metabolism in hypoxic cells is not well understood. Here we show that human cells use reductive metabolism of α-ketoglutarate to synthesize AcCoA for lipid synthesis. This isocitrate dehydrogenase-1 (IDH1)-dependent pathway is active in most cell lines under normal culture conditions, but cells grown under hypoxia rely almost exclusively on the reductive carboxylation of glutamine-derived α-ketoglutarate for de novo lipogenesis. Furthermore, renal cell lines deficient in the von Hippel–Lindau tumour suppressor protein preferentially use reductive glutamine metabolism for lipid biosynthesis even at normal oxygen levels. These results identify a critical role for oxygen in regulating carbon use to produce AcCoA and support lipid synthesis in mammalian cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hatzivassiliou, G. et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 8, 311–321 (2005)
Wellen, K. E. et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009)
Migita, T. et al. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 68, 8547–8554 (2008)
Semenza, G. L. HIF-1: upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 20, 51–56 (2010)
DeBerardinis, R. J. et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl Acad. Sci. USA 104, 19345–19350 (2007)
Weinberg, F. et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl Acad. Sci. USA 107, 8788–8793 (2010)
Des Rosiers, C. et al. Isotopomer analysis of citric acid cycle and gluconeogenesis in rat liver. Reversibility of isocitrate dehydrogenase and involvement of ATP-citrate lyase in gluconeogenesis. J. Biol. Chem. 270, 10027–10036 (1995)
Yoo, H., Antoniewicz, M. R., Stephanopoulos, G. & Kelleher, J. K. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J. Biol. Chem. 283, 20621–20627 (2008)
Metallo, C. M., Walther, J. L. & Stephanopoulos, G. Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J. Biotechnol. 144, 167–174 (2009)
Lemons, J. M. et al. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 8, e1000514 (2010)
Ward, P. S. et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 (2010)
Boros, L. G. et al. Defective RNA ribose synthesis in fibroblasts from patients with thiamine-responsive megaloblastic anemia (TRMA). Blood 102, 3556–3561 (2003)
Munger, J. et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nature Biotechnol. 26, 1179–1186 (2008)
Maier, K., Hofmann, U., Reuss, M. & Mauch, K. Identification of metabolic fluxes in hepatic cells from transient 13C-labeling experiments: part II. Flux estimation. Biotechnol. Bioeng. 100, 355–370 (2008)
Kharroubi, A. T., Masterson, T. M., Aldaghlas, T. A., Kennedy, K. A. & Kelleher, J. K. Isotopomer spectral analysis of triglyceride fatty acid synthesis in 3T3–L1 cells. Am. J. Physiol. 263, E667–E675 (1992)
Siebert, G., Carsiotis, M. & Plaut, G. W. The enzymatic properties of isocitric dehydrogenase. J. Biol. Chem. 226, 977–991 (1957)
Sauer, U. Metabolic networks in motion: 13C-based flux analysis. Mol. Syst. Biol. 2, 62 (2006)
Young, J. D., Walther, J. L., Antoniewicz, M. R., Yoo, H. & Stephanopoulos, G. An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnol. Bioeng. 99, 686–699 (2008)
Hartong, D. T. et al. Insights from retinitis pigmentosa into the roles of isocitrate dehydrogenases in the Krebs cycle. Nature Genet. 40, 1230–1234 (2008)
Cheng, T. et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl Acad. Sci. USA 108, 8674–8679 (2011)
Bonnet, S. et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell 11, 37–51 (2007)
Kim, J. W., Tchernyshyov, I., Semenza, G. L. & Dang, C. V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 (2006)
Papandreou, I., Cairns, R. A., Fontana, L., Lim, A. L. & Denko, N. C. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 3, 187–197 (2006)
Kaelin, W. G., Jr Molecular basis of the VHL hereditary cancer syndrome. Nature Rev. Cancer 2, 673–682 (2002)
Ivan, M. et al. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 (2001)
Jaakkola, P. et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001)
Zimmer, M. et al. Small-molecule inhibitors of HIF-2a translation link its 5′UTR iron-responsive element to oxygen sensing. Mol. Cell 32, 838–848 (2008)
Gordan, J. D., Bertout, J. A., Hu, C. J., Diehl, J. A. & Simon, M. C. HIF-2α promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell 11, 335–347 (2007)
Gao, P. et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765 (2009)
Sutherland, R. M. Cell and environment interactions in tumor microregions: the multicell spheroid model. Science 240, 177–184 (1988)
Zimmer, M., Doucette, D., Siddiqui, N. & Iliopoulos, O. Inhibition of hypoxia-inducible factor is sufficient for growth suppression of VHL−/− tumors. Mol. Cancer Res. 2, 89–95 (2004)
Iliopoulos, O., Kibel, A., Gray, S. & Kaelin, W. G., Jr Tumour suppression by the human von Hippel-Lindau gene product. Nature Med. 1, 822–826 (1995)
Hogquist, K. A. et al. T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994)
Sarbassov, D. D., Guertin, D. A., Ali, S. M. & Sabatini, D. M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 (2005)
Fernandez, C. A., Des Rosiers, C., Previs, S. F., David, F. & Brunengraber, H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J. Mass Spectrom. 31, 255–262 (1996)
Antoniewicz, M. R., Kelleher, J. K. & Stephanopoulos, G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab. Eng. 8, 324–337 (2006)
Antoniewicz, M. R., Kelleher, J. K. & Stephanopoulos, G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab. Eng. 9, 68–86 (2007)
Noguchi, Y. et al. Effect of anaplerotic fluxes and amino acid availability on hepatic lipoapoptosis. J. Biol. Chem. 284, 33425–33436 (2009)
Gaglio, D. et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol. Syst. Biol. 7, 523 (2011)
Grassian, A. R., Metallo, C. M., Coloff, J. L., Stephanopoulos, G. & Brugge, J. S. Erk regulation of pyruvate dehydrogenase flux through PDK4 modulates cell proliferation. Genes Dev. 25, 1716–1733 (2011)
Acknowledgements
We thank N. Vokes and P. Ward for discussions. We also thank S. Gross and Agios Pharmaceuticals for providing the IDH1 construct. We acknowledge support from National Institutes of Health grant R01 DK075850-01. C.M.M. is supported by a postdoctoral fellowship from the American Cancer Society. K.H. is supported by the German Research Foundation (DFG) grant HI1400. L.G. is supported by the NIH and the Glenn Foundation for Medical Research. M.G.V.H. is supported by the Burrough’s Wellcome Fund, the Smith Family, the Damon Runyon Cancer Research Foundation and the National Cancer Institute. D.J.I. is an investigator of the Howard Hughes Medical Institute. O.I. is supported by R01 CA122591 and the Dana Farber/Harvard Cancer Center Kidney SPORE Grant Developmental Award.
Author information
Authors and Affiliations
Contributions
C.M.M., P.A.G., E.L.B., K.R.M., J.Y., K.H. and C.M.J. performed cellular experiments and isotope tracing. C.M.M. and P.A.G. performed metabolite profiling and analysed data. K.R.M. and M.G.V.H. performed enzyme assays and 14C experiments. E.L.B., J.Y. and Z.R.J. generated western blots. D.J.I. and L.G. provided support and reagents. J.K.K., M.G.V.H., O.I. and G.S. provided conceptual advice. C.M.M., J.K.K., M.G.V.H., O.I. and G.S. wrote and edited the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-23 with legends, Supplementary Text on the subject of Metabolic flux analysis (MFA), Supplementary Tables 1-14 and additional references. (PDF 1175 kb)
Rights and permissions
About this article
Cite this article
Metallo, C., Gameiro, P., Bell, E. et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 (2012). https://doi.org/10.1038/nature10602
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10602
This article is cited by
-
Glutamine addiction in tumor cell: oncogene regulation and clinical treatment
Cell Communication and Signaling (2024)
-
Peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) family in physiological and pathophysiological process and diseases
Signal Transduction and Targeted Therapy (2024)
-
Bayesian kinetic modeling for tracer-based metabolomic data
BMC Bioinformatics (2023)
-
Breast cancer cells that preferentially metastasize to lung or bone are more glycolytic, synthesize serine at greater rates, and consume less ATP and NADPH than parent MDA-MB-231 cells
Cancer & Metabolism (2023)
-
Small extracellular vesicle-mediated metabolic reprogramming: from tumors to pre-metastatic niche formation
Cell Communication and Signaling (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.