Abstract
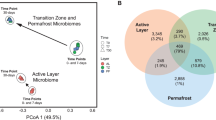
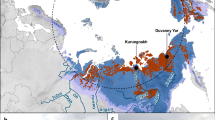
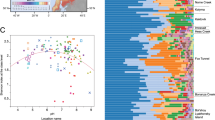
Permafrost contains an estimated 1672 Pg carbon (C), an amount roughly equivalent to the total currently contained within land plants and the atmosphere1,2,3. This reservoir of C is vulnerable to decomposition as rising global temperatures cause the permafrost to thaw2. During thaw, trapped organic matter may become more accessible for microbial degradation and result in greenhouse gas emissions4,5. Despite recent advances in the use of molecular tools to study permafrost microbial communities6,7,8,9, their response to thaw remains unclear. Here we use deep metagenomic sequencing to determine the impact of thaw on microbial phylogenetic and functional genes, and relate these data to measurements of methane emissions. Metagenomics, the direct sequencing of DNA from the environment, allows the examination of whole biochemical pathways and associated processes, as opposed to individual pieces of the metabolic puzzle. Our metagenome analyses reveal that during transition from a frozen to a thawed state there are rapid shifts in many microbial, phylogenetic and functional gene abundances and pathways. After one week of incubation at 5 °C, permafrost metagenomes converge to be more similar to each other than while they are frozen. We find that multiple genes involved in cycling of C and nitrogen shift rapidly during thaw. We also construct the first draft genome from a complex soil metagenome, which corresponds to a novel methanogen. Methane previously accumulated in permafrost is released during thaw and subsequently consumed by methanotrophic bacteria. Together these data point towards the importance of rapid cycling of methane and nitrogen in thawing permafrost.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Schuur, E. A. G. et al. Vulnerability of permafrost carbon to climate change: Implications for the global carbon cycle. Bioscience 58, 701–714 (2008)
Tarnocai, C. et al. Soil organic carbon pools in the northern circumpolar permafrost region. Glob. Biogeochem. Cycles 23, GB2023 (2009)
Zimov, S. A., Schuur, E. A. & Chapin, F. S. Climate change. Permafrost and the global carbon budget. Science 312, 1612–1613 (2006)
Osterkamp, T. Characteristics of the recent warming of permafrost in Alaska. J. Geophys. Res. 112, F02S02 (2007)
Prater, J. L., Chanton, J. P. & Whiting, G. J. Variation in methane production pathways associated with permafrost decomposition in collapse scar bogs of Alberta, Canada. Glob. Biogeochem. Cycles 21, GB4004 (2007)
Chu, H. et al. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 12, 2998–3006 (2010)
Hansen, A. A. et al. Viability, diversity and composition of the bacterial community in a high Arctic permafrost soil from Spitsbergen, Northern Norway. Environ. Microbiol. 9, 2870–2884 (2007)
Steven, B., Pollard, W. H., Greer, C. W. & Whyte, L. G. Microbial diversity and activity through a permafrost/ground ice core profile from the Canadian high Arctic. Environ. Microbiol. 10, 3388–3403 (2008)
Yergeau, E., Hogues, H., Whyte, L. G. & Greer, C. W. The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR and microarray analyses. ISME J. 4, 1206–1214 (2010)
Waldrop, M. P. et al. Molecular investigations into a globally important carbon pool: permafrost-protected carbon in Alaskan soils. Glob. Change Biol. 16, 2543–2544 (2010)
Rivkina, R. et al. Biogeochemistry of methane and methanogenic archaea in permafrost. FEMS Microbiol. Ecol. 61, 1–15 (2007)
Trotsenko, Y. A. & Khmelenian, V. N. Aerobic methanotrophic bacteria of cold ecosystems. FEMS Microbiol. Ecol. 53, 15–26 (2005)
Blow, M. et al. Identification of ancient remains through genomic sequencing. Genome Res. 18, 1347–1353 (2008)
Hess, M. et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331, 463–467 (2011)
Sakai, S. et al. Methanocella paludicola gen. nov., sp. nov., a methane-producing archaeon, the first isolate of the lineage ‘Rice Cluster I’, and proposal of the new archaeal order Methanocellales ord. nov. Int. J. Syst. Evol. Microbiol. 58, 929–936 (2008)
Murray, P. A. & Zinder, S. H. Nitrogen-fixation by a methanogenic archaebacterium. Nature 312, 284–286 (1984)
Johnson, S. S. et al. Ancient bacteria show evidence of DNA repair. Proc. Natl Acad. Sci. USA 104, 14401–14405 (2007)
Ferry, J. G. How to make a living by exhaling methane. Annu. Rev. Microbiol. 64, 453–473 (2010)
Williams, R. et al. Amplification of complex gene libraries by emulsion PCR. Nature Methods 3, 545–550 (2006)
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000)
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990)
DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006)
Pruesse, E. C. et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196 (2007)
Zerbino, D. R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008)
Markowitz, V. M. et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 36, D534–D538 (2008)
DeAngelis, K. M. et al. Selective progressive response of soil microbial community to wild oat roots. ISME J. 3, 168–178 (2009)
Brodie, E., Edwards, S. & Clipson, N. Bacterial community dynamics across a floristic gradient in a temperate upland grassland ecosystem. Microb. Ecol. 44, 260–270 (2002)
Griffiths, R. I., Whiteley, A. S., O’Donnell, A. G. & Bailey, M. J. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66, 5488–5491 (2000)
Kunin, V., Engelbrektson, A., Ochman, H. & Hugenholtz, P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 12, 118–123 (2010)
Kunin, V. & Hugenholtz, P. PyroTagger: a fast, accurate pipeline for analysis of rRNA amplicon pyrosequence data. Open J. 1, 1 (2010)
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010)
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009)
Hamady, M., Lozupone, C. & Knight, R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4, 17–27 (2010)
White, J. R., Nagarajan, N. & Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 5, e1000352 (2009)
R Development Core Team . R: A Language and Environment for Statistical Computing 〈http://www.R-project.org〉 (2010)
Kristiansson, E., Hugenholtz, P. & Dalevi, D. ShotgunFunctionalizeR: an R-package for functional comparison of metagenomes. Bioinformatics 25, 2737–2738 (2009)
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J.R. Stat. Soc. B 57, 289–300 (1995)
Huson, D. H., Auch, A. F., Qi, J. & Schuster, S. C. MEGAN analysis of metagenomic data. Genome Res. 17, 377–386 (2007)
Oksanen, J. et al. Vegan: Community Ecology Package. R package version 1.15-4. 〈http://CRAN.R-project.org/package=vegan〉 (2011)
Suzuki, R. & Shimodaira, H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Wu, M. & Eisen, J. A. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 9, R151 (2008)
Talavera, G. & Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignment. Syst. Biol. 56, 564–577 (2007)
David, M. M., Sapkota, A. R., Simonet, P. & Vogel, T. M. A novel and rapid method for synthesizing positive controls and standards for quantitative PCR. J. Microbiol. Methods 73, 73–77 (2008)
Acknowledgements
The work conducted by the Lawrence Berkeley National Laboratory Earth Sciences Division (Laboratory Directed Research Development) and the Joint Genome Institute was supported in part by the Office of Science of the US Department of Energy under Contract no. DE-AC02-05CH11231. This study was also supported by the Venture Capital and Yukon River Basin project of the United States Geological Survey. We acknowledge the technical support by the Joint Genome Institute production team. We thank A. Sczyrba, R. Egan and S. Canon for discussions and advice.
Author information
Authors and Affiliations
Contributions
J.K.J., M.P.W. and K.M.D. conceived the incubation experiments. M.P.W. collected the samples. M.P.W. and S.J.B. conducted the incubation experiments. R.M., K.L.C. and K.M.D. performed the DNA extractions. R.M. created the shotgun sequencing libraries and conducted bioinformatics analyses. R.M. and K.M.D. performed statistical analyses. R.M. and M.M.D. performed qPCR experiments. R.M. and J.K.J. wrote the paper. All authors discussed the results and commented on the manuscript. E.M.R. M.P.W. and J.K.J. obtained funding for the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
All annotated assembled sequences were incorporated into the IMG/M system with the Taxon Object ID 2067725009. Raw Illumina and 454 pyrotag sequence reads and a list containing the subset of contigs belonging to the draft methanogen genome are available at https://www.jgi.doe.gov/downloads/Permafrost_metagenome.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-16 with legends, Supplementary Tables 1-7 and additional references. (PDF 4748 kb)
Rights and permissions
About this article
Cite this article
Mackelprang, R., Waldrop, M., DeAngelis, K. et al. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480, 368–371 (2011). https://doi.org/10.1038/nature10576
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10576
This article is cited by
-
Assessing the microbiota of the snail intermediate host of trematodes, Galba truncatula
Parasites & Vectors (2024)
-
Distinct taxonomic and functional profiles of high Arctic and alpine permafrost-affected soil microbiomes
Environmental Microbiome (2023)
-
Warming changes the composition and diversity of fungal communities in permafrost
Annals of Microbiology (2023)
-
Hot moment of N2O emissions in seasonally frozen peatlands
The ISME Journal (2023)
-
Extreme summers impact cropland and grassland soil microbiomes
The ISME Journal (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.