Abstract
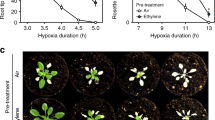
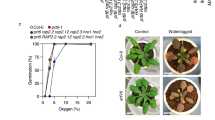
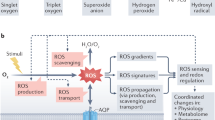
The majority of eukaryotic organisms rely on molecular oxygen for respiratory energy production1. When the supply of oxygen is compromised, a variety of acclimation responses are activated to reduce the detrimental effects of energy depletion2,3,4. Various oxygen-sensing mechanisms have been described that are thought to trigger these responses5,6,7,8,9, but they each seem to be kingdom specific and no sensing mechanism has been identified in plants until now. Here we show that one branch of the ubiquitin-dependent N-end rule pathway for protein degradation, which is active in both mammals and plants10,11, functions as an oxygen-sensing mechanism in Arabidopsis thaliana. We identified a conserved amino-terminal amino acid sequence of the ethylene response factor (ERF)-transcription factor RAP2.12 to be dedicated to an oxygen-dependent sequence of post-translational modifications, which ultimately lead to degradation of RAP2.12 under aerobic conditions. When the oxygen concentration is low—as during flooding—RAP2.12 is released from the plasma membrane and accumulates in the nucleus to activate gene expression for hypoxia acclimation. Our discovery of an oxygen-sensing mechanism opens up new possibilities for improving flooding tolerance in crops.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
The raw data files of the microarray experiments have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo; accession number: GSE29187). The gene sequences for the Rumex spp. used in this work have been deposited at NCBI (RaERF1: JF968115; RaERF2: JF968116; RpERF1: JF968117; RpERF2: JF968118; and RpERF3: JF968119).
References
Webb, J. D., Coleman, M. L. & Pugh, C. W. Hypoxia, hypoxia-inducible factors (HIF), HIF hydroxylases and oxygen sensing. Cell. Mol. Life Sci. 66, 3539–3554 (2009)
Kelly, D. P. Hypoxic reprogramming. Nature Genet. 40, 132–134 (2008)
Mustroph, A. et al. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 152, 1484–1500 (2010)
Bailey-Serres, J. & Voesenek, L. A. C. J. Flooding stress: acclimations and genetic diversity. Annu. Rev. Plant Biol. 59, 313–339 (2008)
Green, J., Crack, J. C., Thomson, A. J. & LeBrun, N. E. Bacterial sensors of oxygen. Curr. Opin. Microbiol. 12, 145–151 (2009)
Hou, S. et al. Myoglobin-like aerotaxis transducers in Archaea and Bacteria. Nature 403, 540–544 (2000)
Osborne, T. F. & Espenshade, P. J. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it’s been. Genes Dev. 23, 2578–2591 (2009)
Semenza, G. L. HIF-1, O2, and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell 107, 1–3 (2001)
van der Wel, H. et al. Requirements for Skp1 processing by cytosolic Prolyl 4(trans)-hydroxylase and α-N-acetylglucosaminyltransferase enzymes involved in O2 signaling in Dictyostelium . Biochemistry 50, 1700–1713 (2011)
Lee, M. J. et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl Acad. Sci. USA 102, 15030–15035 (2005)
Graciet, E., Mesiti, F. & Wellmer, F. Structure and evolutionary conservation of the plant N-end rule pathway. Plant J. 61, 741–751 (2010)
Hinz, M. et al. Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 153, 757–772 (2010)
Licausi, F. et al. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana . Plant J. 62, 302–315 (2010)
Xu, K. et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708 (2006)
Hattori, Y. et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030 (2009)
Papdi, C. et al. Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol. 147, 528–542 (2008)
Licausi, F. et al. Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytol. 190, 442–456 (2011)
Li, H. Y. & Chye, M. L. Membrane localization of Arabidopsis acyl-CoA binding protein ACBP2. Plant Mol. Biol. 51, 483–492 (2003)
Li, H. Y. & Chye, M. L. Arabidopsis Acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element-binding protein, AtEBP, via its ankyrin repeats. Plant Mol. Biol. 54, 233–243 (2004)
Okamuro, J. K., Caster, B., Villarroel, R., Van Montagu, M. & Jofuku, K. D. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis . Proc. Natl Acad. Sci. USA 94, 7076–7081 (1997)
Kwon, Y. T. et al. An essential role of N-terminal arginylation in cardiovascular development. Science 297, 96–99 (2002)
Graciet, E. & Wellmer, F. The plant N-end rule pathway: structure and functions. Trends Plant Sci. 15, 447–453 (2010)
Bradshaw, R. A., Brickey, W. W. & Walker, K. W. N-terminal processing: the methionine aminopeptidase and Nα-acetyl transferase families. Trends Biochem. Sci. 23, 263–267 (1998)
Garzón, M. et al. PRT6/At5g02310 encodes an Arabidopsis ubiquitin ligase of the N-end rule pathway with arginine specificity and is not the CER3 locus. FEBS Lett. 581, 3189–3196 (2007)
Voges, D., Zwickl, P. & Baumeister, W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068 (2000)
Vallon, U. & Kull, U. Localization of proteasomes in plant cells. Protoplasma 182, 15–18 (1994)
Pierik, R., de Wit, M. & Voesenek, L. A. C. J. Growth-mediated stress escape: convergence of signal transduction pathways activated upon exposure to two different environmental stresses. New Phytol. 189, 122–134 (2011)
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W. & Chua, N. H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nature Protocols 1, 641–646 (2006)
Lee, M. W. & Yang, Y. Transient expression assay by agroinfiltration of leaves. Methods Mol. Biol. 323, 225–229 (2006)
Lohse, M. et al. Robin: An intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol. 153, 642–651 (2010)
Smyth, G. K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article 3. (2004)
Reiner, A., Yekutieli, D. & Benjamini, Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375 (2003)
Wu, Z., Irizarry, R. A., Gentleman, R., Martinez-Murillo, F. & Spencer, F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99, 909–917 (2004)
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572 (2007)
Gehl, C., Waadt, R., Kudla, J., Mendel, R.-R. & Hänsch, R. New GATEWAY vectors for high throughput analyses of protein–protein interactions by bimolecular fluorescence complementation. Mol. Plant 2, 1051–1058 (2009)
Acknowledgements
We would like to thank H. van Veen and R. Sasidharan (for providing Rumex data), E. Maximova, F. Kragler (microscopy), W. Schulze and R. Bock (support and discussion), A. Fernie and R. Pierik (commenting on the manuscript) and S. Parlanti, L. Bartezko and K. Seehaus (plant cultivation). This work was financially supported by the Max Planck Institute of Molecular Plant Physiology, Scuola Superiore Sant’Anna, and the Deutsche Forschungsgemeinschaft (DFG) (DO 1298/2-1).
Author information
Authors and Affiliations
Contributions
F.L., M.K., D.A.W. and B.G. performed the experiments. F.M.G. carried out the bioinformatical analysis. F.L., L.A.C.J.V., P.P. and J.T.v.D. designed the experiments. F.L., P.P. and J.T.v.D. wrote the manuscript. All the authors discussed and commented on the content of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-16 with legends, Supplementary Tables 1-8 and additional references. (PDF 3312 kb)
Rights and permissions
About this article
Cite this article
Licausi, F., Kosmacz, M., Weits, D. et al. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422 (2011). https://doi.org/10.1038/nature10536
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10536
This article is cited by
-
Candidate Gene Transcriptional Signature Unravels the Reprogramming Occurring in the Peel of Apple Fruit of ‘Granny Smith’ During Postharvest Storage
Journal of Plant Growth Regulation (2024)
-
Navigating Through Harsh Conditions: Coordinated Networks of Plant Adaptation to Abiotic Stress
Journal of Plant Growth Regulation (2024)
-
Histological, physio-biochemical, and transcriptomic analyses reveal the potential limiting factors for successful grafting of pecan
Journal of Forestry Research (2024)
-
Iron-sulfur clusters are involved in post-translational arginylation
Nature Communications (2023)
-
ERFVII action and modulation through oxygen-sensing in Arabidopsis thaliana
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.