Abstract
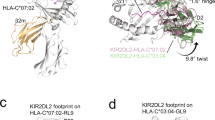
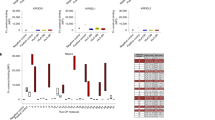
Members of the killer cell immunoglobulin-like receptor (KIR) family, a large group of polymorphic receptors expressed on natural killer (NK) cells, recognize particular peptide-laden human leukocyte antigen (pHLA) class I molecules and have a pivotal role in innate immune responses1. Allelic variation and extensive polymorphism within the three-domain KIR family (KIR3D, domains D0–D1–D2) affects pHLA binding specificity and is linked to the control of viral replication and the treatment outcome of certain haematological malignancies1,2,3. Here we describe the structure of a human KIR3DL1 receptor bound to HLA-B*5701 complexed with a self-peptide. KIR3DL1 clamped around the carboxy-terminal end of the HLA-B*5701 antigen-binding cleft, resulting in two discontinuous footprints on the pHLA. First, the D0 domain, a distinguishing feature of the KIR3D family, extended towards β2-microglobulin and abutted a region of the HLA molecule with limited polymorphism, thereby acting as an ‘innate HLA sensor’ domain. Second, whereas the D2–HLA-B*5701 interface exhibited a high degree of complementarity, the D1–pHLA-B*5701 contacts were suboptimal and accommodated a degree of sequence variation both within the peptide and the polymorphic region of the HLA molecule. Although the two-domain KIR (KIR2D) and KIR3DL1 docked similarly onto HLA-C4,5 and HLA-B respectively, the corresponding D1-mediated interactions differed markedly, thereby providing insight into the specificity of KIR3DL1 for discrete HLA-A and HLA-B allotypes. Collectively, in association with extensive mutagenesis studies at the KIR3DL1–pHLA-B*5701 interface, we provide a framework for understanding the intricate interplay between peptide variability, KIR3D and HLA polymorphism in determining the specificity requirements of this essential innate interaction that is conserved across primate species.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Parham, P. MHC class I molecules and KIRs in human history, health and survival. Nature Rev. Immunol. 5, 201–214 (2005)
Sanjanwala, B., Draghi, M., Norman, P. J., Guethlein, L. A. & Parham, P. Polymorphic sites away from the Bw4 epitope that affect interaction of Bw4+ HLA-B with KIR3DL1. J. Immunol. 181, 6293–6300 (2008)
Martin, M. P. et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nature Genet. 39, 733–740 (2007)
Boyington, J. C., Motyka, S. A., Schuck, P., Brooks, A. G. & Sun, P. D. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature 405, 537–543 (2000)
Fan, Q. R., Long, E. O. & Wiley, D. C. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1–HLA–Cw4 complex. Nature Immunol. 2, 452–460 (2001)
Colonna, M. & Samaridis, J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science 268, 405–408 (1995)
Chessman, D. et al. Human leukocyte antigen class I-restricted activation of CD8+ T cells provides the immunogenetic basis of a systemic drug hypersensitivity. Immunity 28, 822–832 (2008)
Litwin, V., Gumperz, J., Parham, P., Phillips, J. H. & Lanier, L. L. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J. Exp. Med. 180, 537–543 (1994)
Gumperz, J. E., Litwin, V., Phillips, J. H., Lanier, L. L. & Parham, P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J. Exp. Med. 181, 1133–1144 (1995)
Cella, M., Longo, A., Ferrara, G. B., Strominger, J. L. & Colonna, M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J. Exp. Med. 180, 1235–1242 (1994)
Petrie, E. J. et al. CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence. J. Exp. Med. 205, 725–735 (2008)
Hülsmeyer, M. et al. Thermodynamic and structural equivalence of two HLA-B27 subtypes complexed with a self-peptide. J. Mol. Biol. 346, 1367–1379 (2005)
Peruzzi, M., Parker, K. C., Long, E. O. & Malnati, M. S. Peptide sequence requirements for the recognition of HLA-B*2705 by specific natural killer cells. J. Immunol. 157, 3350–3356 (1996)
Fadda, L. et al. Peptide antagonism as a mechanism for NK cell activation. Proc. Natl Acad. Sci. USA 107, 10160–10165 (2010)
Maenaka, K. et al. Killer cell immunoglobulin receptors and T cell receptors bind peptide-major histocompatibility complex class I with distinct thermodynamic and kinetic properties. J. Biol. Chem. 274, 28329–28334 (1999)
Trowsdale, J. et al. The genomic context of natural killer receptor extended gene families. Immunol. Rev. 181, 20–38 (2001)
Norman, P. J. et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nature Genet. 39, 1092–1099 (2007)
Martin, M. P. et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nature Genet. 31, 429–434 (2002)
Carr, W. H. et al. Cutting edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J. Immunol. 178, 647–651 (2007)
Sharma, D. et al. Dimorphic motifs in D0 and D1+D2 domains of killer cell Ig-like receptor 3DL1 combine to form receptors with high, moderate, and no avidity for the complex of a peptide derived from HIV and HLA-A*2402. J. Immunol. 183, 4569–4582 (2009)
O’Connor, G. M. et al. Analysis of binding of KIR3DS1*014 to HLA suggests distinct evolutionary history of KIR3DS1. J. Immunol. 187, 2162–2171 (2011)
Dohring, C., Scheidegger, D., Samaridis, J., Cella, M. & Colonna, M. A human killer inhibitory receptor specific for HLA-A1,2. J. Immunol. 156, 3098–3101 (1996)
Hansasuta, P. et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur. J. Immunol. 34, 1673–1679 (2004)
Yawata, M. et al. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 203, 633–645 (2006)
Khakoo, S. I., Geller, R., Shin, S., Jenkins, J. A. & Parham, P. The D0 domain of KIR3D acts as a major histocompatibility complex class I binding enhancer. J. Exp. Med. 196, 911–921 (2002)
Carr, W. H., Pando, M. J. & Parham, P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J. Immunol. 175, 5222–5229 (2005)
Godfrey, D. I., Rossjohn, J. & McCluskey, J. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity 28, 304–314 (2008)
Aricescu, A. R., Lu, W. & Jones, E. Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D 62, 1243–1250 (2006)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
Acknowledgements
We thank the staff at the MX2 beamline of the Australian synchrotron for assistance with data collection. We thank A. Radu Aricescu for the gift of the pHLsec vector. This research was supported by the National Health and Medical Research Council of Australia (NHMRC), the Australian Research Council (ARC) and the Intramural Research Programs of the National Cancer Institute and the National Institute of Allergy and Infectious Diseases, National Institutes of Health. D.W.M. and G.M.O'C. were supported by the Intramural AIDS Targeted Antiviral Program of the National Institutes of Health. J.P.V. is supported by an NHMRC Peter Doherty Research Fellowship; D.A.P. is supported by a Medical Research Council (UK) Senior Clinical Fellowship; B.A.P.L. is supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health; C.S.C. is supported by an ARC QEII Fellowship; J.R. is supported by an ARC Federation Fellowship.
Author information
Authors and Affiliations
Contributions
J.P.V. solved the structure, undertook analysis, performed experiments and contributed to manuscript preparation. H.H.R., T.B., R.C.D., R.B., P.M.S., M.A.O., J.M.L.W., C.M.H., J.L., S.M.M., S.G. and C.S.C. performed experiments and/or analysed data. G.M.O’C., D.A.P., B.A.P.L. and D.W.M. performed experiments and/or analysed data and contributed to the writing of the manuscript; A.G.B. and J.R. were the joint senior authors—they co-led the investigation, devised the project, analysed the data and wrote the manuscript.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-7 with legends and Supplementary Tables 1-3. (PDF 15308 kb)
Rights and permissions
About this article
Cite this article
Vivian, J., Duncan, R., Berry, R. et al. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature 479, 401–405 (2011). https://doi.org/10.1038/nature10517
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10517
This article is cited by
-
HLA-B*44 and the Bw4-80T motif are associated with poor outcome of relapse-preventive immunotherapy in acute myeloid leukemia
Cancer Immunology, Immunotherapy (2023)
-
Structural plasticity of KIR2DL2 and KIR2DL3 enables altered docking geometries atop HLA-C
Nature Communications (2021)
-
Modulation of innate and adaptive immunity by cytomegaloviruses
Nature Reviews Immunology (2020)
-
A subset of HLA-DP molecules serve as ligands for the natural cytotoxicity receptor NKp44
Nature Immunology (2019)
-
Adverse drug reactions triggered by the common HLA-B*57:01 variant: virtual screening of DrugBank using 3D molecular docking
Journal of Cheminformatics (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.