Abstract
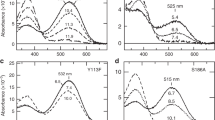
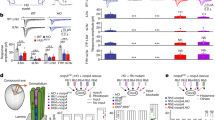
Sensory systems with high discriminatory power use neurons that express only one of several alternative sensory receptor proteins. This exclusive receptor gene expression restricts the sensitivity spectrum of neurons and is coordinated with the choice of their synaptic targets1,2,3. However, little is known about how it is maintained throughout the life of a neuron. Here we show that the green-light sensing receptor rhodopsin 6 (Rh6) acts to exclude an alternative blue-sensitive rhodopsin 5 (Rh5) from a subset of Drosophila R8 photoreceptor neurons4. Loss of Rh6 leads to a gradual expansion of Rh5 expression into all R8 photoreceptors of the ageing adult retina. The Rh6 feedback signal results in repression of the rh5 promoter and can be mimicked by other Drosophila rhodopsins; it is partly dependent on activation of rhodopsin by light, and relies on Gαq activity, but not on the subsequent steps of the phototransduction cascade5. Our observations reveal a thus far unappreciated spectral plasticity of R8 photoreceptors, and identify rhodopsin feedback as an exclusion mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mombaerts, P. Axonal wiring in the mouse olfactory system. Annu. Rev. Cell Dev. Biol. 22, 713–737 (2006)
Komiyama, T. & Luo, L. Development of wiring specificity in the olfactory system. Curr. Opin. Neurobiol. 16, 67–73 (2006)
Morey, M. et al. Coordinate control of synaptic-layer specificity and rhodopsins in photoreceptor neurons. Nature 456, 795–799 (2008)
Rister, J. & Desplan, C. The retinal mosaics of opsin expression in invertebrates and vertebrates. Dev. Neurobiol. 10.1002/dneu.20905 (2011)
Wang, T. & Montell, C. Phototransduction and retinal degeneration in Drosophila . Pflügers Arch. 454, 821–847 (2007)
Franceschini, N., Kirschfeld, K. & Minke, B. Fluorescence of photoreceptor cells observed in vivo . Science 213, 1264–1267 (1981)
Mazzoni, E. O. et al. Iroquois complex genes induce co-expression of rhodopsins in Drosophila . PLoS Biol. 6, e97 (2008)
Mikeladze-Dvali, T. et al. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell 122, 775–787 (2005)
Wernet, M. F. et al. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature 440, 174–180 (2006)
Serizawa, S. et al. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science 302, 2088–2094 (2003)
Shykind, B. M. et al. Gene switching and the stability of odorant receptor gene choice. Cell 117, 801–815 (2004)
Lewcock, J. W. & Reed, R. R. A feedback mechanism regulates monoallelic odorant receptor expression. Proc. Natl Acad. Sci. USA 101, 1069–1074 (2004)
Feinstein, P., Bozza, T., Rodriguez, I., Vassalli, A. & Mombaerts, P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the β2 adrenergic receptor. Cell 117, 833–846 (2004)
Fuss, S. H. & Ray, A. Mechanisms of odorant receptor gene choice in Drosophila and vertebrates. Mol. Cell. Neurosci. 41, 101–112 (2009)
Cook, T., Pichaud, F., Sonneville, R., Papatsenko, D. & Desplan, C. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila . Dev. Cell 4, 853–864 (2003)
Harris, W. A., Stark, W. S. & Walker, J. A. Genetic dissection of the photoreceptor system in the compound eye of Drosophila melanogaster . J. Physiol. (Lond.) 256, 415–439 (1976)
Chou, W. H. et al. Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development 126, 607–616 (1999)
Papatsenko, D., Sheng, G. & Desplan, C. A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development 124, 1665–1673 (1997)
Chou, W. H. et al. Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron 17, 1101–1115 (1996)
Tahayato, A. et al. Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev. Cell 5, 391–402 (2003)
Green, P., Hartenstein, A. Y. & Hartenstein, V. The embryonic development of the Drosophila visual system. Cell Tissue Res. 273, 583–598 (1993)
Sprecher, S. G., Pichaud, F. & Desplan, C. Adult and larval photoreceptors use different mechanisms to specify the same Rhodopsin fates. Genes Dev. 21, 2182–2195 (2007)
Sprecher, S. G. & Desplan, C. Switch of rhodopsin expression in terminally differentiated Drosophila sensory neurons. Nature 454, 533–537 (2008)
Hofbauer, A. & Buchner, E. Does Drosophila have seven eyes? Naturwissenschaften 76, 335–336 (1989)
Yasuyama, K. & Meinertzhagen, I. A. Extraretinal photoreceptors at the compound eye’s posterior margin in Drosophila melanogaster . J. Comp. Neurol. 412, 193–202 (1999)
Huang, J. et al. Activation of TRP channels by protons and phosphoinositide depletion in Drosophila photoreceptors. Curr. Biol. 20, 189–197 (2010)
O’Tousa, J. E., Leonard, D. S. & Pak, W. L. Morphological defects in oraJK84 photoreceptors caused by mutation in R1–6 opsin gene of Drosophila . J. Neurogenet. 6, 41–52 (1989)
Kumar, J. P. & Ready, D. F. Rhodopsin plays an essential structural role in Drosophila photoreceptor development. Development 121, 4359–4370 (1995)
Shen, W. L. et al. Function of rhodopsin in temperature discrimination in Drosophila . Science 331, 1333–1336 (2011)
Arikawa, K., Mizuno, S., Kinoshita, M. & Stavenga, D. G. Coexpression of two visual pigments in a photoreceptor causes an abnormally broad spectral sensitivity in the eye of the butterfly Papilio xuthus . J. Neurosci. 23, 4527–4532 (2003)
Earl, J. B. & Britt, S. G. Expression of Drosophila rhodopsins during photoreceptor cell differentiation: insights into R7 and R8 cell subtype commitment. Gene Expr. Patterns 6, 687–694 (2006)
Parks, A. L. et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nature Genet. 36, 288–292 (2004)
Oberstein, A., Pare, A., Kaplan, L. & Small, S. Site-specific transgenesis by Cre-mediated recombination in Drosophila . Nature Methods 2, 583–585 (2005)
Dietzl, G. et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila . Nature 448, 151–156 (2007)
Till, B. J. et al. High-throughput TILLING for functional genomics. Methods Mol. Biol. 236, 205–220 (2003)
Lai, S. L. & Lee, T. Genetic mosaic with dual binary transcriptional systems in Drosophila . Nature Neurosci. 9, 703–709 (2006)
Stapleton, M. et al. The Drosophila gene collection: identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 12, 1294–1300 (2002)
Bischof, J., Maeda, R. K., Hediger, M., Karch, F. & Basler, K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl Acad. Sci. USA 104, 3312–3317 (2007)
Markstein, M., Pitsouli, C., Villalta, C., Celniker, S. E. & Perrimon, N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nature Genet. 40, 476–483 (2008)
Scott, K., Becker, A., Sun, Y., Hardy, R. & Zuker, C. Gq alpha protein function in vivo: genetic dissection of its role in photoreceptor cell physiology. Neuron 15, 919–927 (1995)
Bloomquist, B. T. et al. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell 54, 723–733 (1988)
Yamaguchi, S., Wolf, R., Desplan, C. & Heisenberg, M. Motion vision is independent of color in Drosophila . Proc. Natl Acad. Sci. USA 105, 4910–4915 (2008)
Gerresheim, F. Isolation of Drosophila melanogaster mutants with a wavelength-specific alteration in their phototactic response. Behav. Genet. 18, 227–246 (1988)
Niemeyer, B. A., Suzuki, E., Scott, K., Jalink, K. & Zuker, C. S. The Drosophila light-activated conductance is composed of the two channels TRP and TRPL. Cell 85, 651–659 (1996)
Papatsenko, D., Nazina, A. & Desplan, C. A conserved regulatory element present in all Drosophila rhodopsin genes mediates Pax6 functions and participates in the fine-tuning of cell-specific expression. Mech. Dev. 101, 143–153 (2001)
Justice, R. W., Zilian, O., Woods, D. F., Noll, M. & Bryant, P. J. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9, 534–546 (1995)
Xu, T., Wang, W., Zhang, S., Stewart, R. A. & Yu, W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053–1063 (1995)
O’Keefe, L. V. et al. Drosophila orthologue of WWOX, the chromosomal fragile site FRA16D tumour suppressor gene, functions in aerobic metabolism and regulates reactive oxygen species. Hum. Mol. Genet. 20, 497–509 (2011)
Acknowledgements
We thank J. Blau, B. Collins, M. Cols, T. Erclik, S.H. Fuss, D. Jukam, J.P. Kumar, E. Laufer, H.-S. Li, B. Minke, C. Montell, F. Pichaud, J. Rister and A. Tomlinson for suggestions and comments on the manuscript, V. Douard for help with qRT–PCR, J. Goodness for help identifying rh6fs allele, S.G. Britt, P.J. Dolph, P.R. Hiesinger, F. Pichaud, N. Pinal, D.F. Ready, C.S. Zuker, and the Bloomington Drosophila Stock Center for flies or antibodies. This work was funded by the National Institutes of Health R01EY13012 to C.D. and F32EY016309 to D.V.
Author information
Authors and Affiliations
Contributions
D.V., E.O.M. and C.D. conceived the experiments; D.V. and E.O.M. designed and performed experiments in adult flies; S.G.S. designed and performed experiments in larvae; K.B. performed RNAi experiments; R.J.J., P.L., N.V. and A.C. contributed reagents; D.V. and C.D. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-9 with legends and Supplementary Tables 1-3. (PDF 7386 kb)
Rights and permissions
About this article
Cite this article
Vasiliauskas, D., Mazzoni, E., Sprecher, S. et al. Feedback from rhodopsin controls rhodopsin exclusion in Drosophila photoreceptors. Nature 479, 108–112 (2011). https://doi.org/10.1038/nature10451
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10451
This article is cited by
-
Neurexin-1-dependent circuit activity is required for the maintenance of photoreceptor subtype identity in Drosophila
Molecular Brain (2024)
-
Dietary restriction and the transcription factor clock delay eye aging to extend lifespan in Drosophila Melanogaster
Nature Communications (2022)
-
Muscarinic acetylcholine receptor signaling generates OFF selectivity in a simple visual circuit
Nature Communications (2019)
-
A rhodopsin in the brain functions in circadian photoentrainment in Drosophila
Nature (2017)
-
Phenotypic plasticity in opsin expression in a butterfly compound eye complements sex role reversal
BMC Evolutionary Biology (2012)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.