Abstract
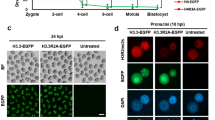
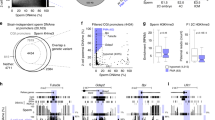
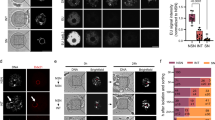
Sperm and eggs carry distinctive epigenetic modifications that are adjusted by reprogramming after fertilization1. The paternal genome in a zygote undergoes active DNA demethylation before the first mitosis2,3. The biological significance and mechanisms of this paternal epigenome remodelling have remained unclear4. Here we report that, within mouse zygotes, oxidation of 5-methylcytosine (5mC) occurs on the paternal genome, changing 5mC into 5-hydroxymethylcytosine (5hmC). Furthermore, we demonstrate that the dioxygenase Tet3 (ref. 5) is enriched specifically in the male pronucleus. In Tet3-deficient zygotes from conditional knockout mice, paternal-genome conversion of 5mC into 5hmC fails to occur and the level of 5mC remains constant. Deficiency of Tet3 also impedes the demethylation process of the paternal Oct4 and Nanog genes and delays the subsequent activation of a paternally derived Oct4 transgene in early embryos. Female mice depleted of Tet3 in the germ line show severely reduced fecundity and their heterozygous mutant offspring lacking maternal Tet3 suffer an increased incidence of developmental failure. Oocytes lacking Tet3 also seem to have a reduced ability to reprogram the injected nuclei from somatic cells. Therefore, Tet3-mediated DNA hydroxylation is involved in epigenetic reprogramming of the zygotic paternal DNA following natural fertilization and may also contribute to somatic cell nuclear reprogramming during animal cloning.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Surani, M. A., Hayashi, K. & Hajkova, P. Genetic and epigenetic regulators of pluripotency. Cell 128, 747–762 (2007)
Mayer, W., Niveleau, A., Walter, J., Fundele, R. & Haaf, T. Demethylation of the zygotic paternal genome. Nature 403, 501–502 (2000)
Oswald, J. et al. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 10, 475–478 (2000)
Ooi, S. K. & Bestor, T. H. The colorful history of active DNA demethylation. Cell 133, 1145–1148 (2008)
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009)
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009)
Iqbal, K., Jin, S. G., Pfeifer, G. P. & Szabo, P. E. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl Acad. Sci. USA 108, 3642–3647 (2011)
Wossidlo, M. et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nature Commun. 2, 241 (2011)
Wu, S. C. & Zhang, Y. Active DNA demethylation: many roads lead to Rome. Nature Rev. Mol. Cell Biol. 11, 607–620 (2010)
Jamil, A. Z., Iqbal, K., Fawad Ur, R. & Mirza, K. A. Effect of phacoemulsification on intraocular pressure. J. Coll. Physicians Surg. Pak. 21, 347–350 (2011)
Wossidlo, M. et al. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 29, 1877–1888 (2010)
Jin, S. G., Kadam, S. & Pfeifer, G. P. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 38, e125 (2010)
Huang, Y. et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE 5, e8888 (2010)
Hattori, N. et al. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 279, 17063–17069 (2004)
Imamura, M. et al. Transcriptional repression and DNA hypermethylation of a small set of ES cell marker genes in male germline stem cells. BMC Dev. Biol. 6, 34 (2006)
Farthing, C. R. et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 4, e1000116 (2008)
Iqbal, K. et al. Subcutaneous panniculitis-like T-cell lymphoma in association with sarcoidosis. Clin. Exp. Dermatol. 36, 677–679 (2011)
Bui, H. T. et al. The cytoplasm of mouse germinal vesicle stage oocytes can enhance somatic cell nuclear reprogramming. Development 135, 3935–3945 (2008)
Yang, H. et al. High-efficiency somatic reprogramming induced by intact MII oocytes. Cell Res. 20, 1034–1042 (2010)
Egli, D., Rosains, J., Birkhoff, G. & Eggan, K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature 447, 679–685 (2007)
Egli, D., Sandler, V. M., Shinohara, M. L., Cantor, H. & Eggan, K. Reprogramming after chromosome transfer into mouse blastomeres. Curr. Biol. 19, 1403–1409 (2009)
Seidman, J. G. & Seidman, C. Transcription factor haploinsufficiency: when half a loaf is not enough. J. Clin. Invest. 109, 451–455 (2002)
Li, L. et al. The mildiomycin biosynthesis: initial steps for sequential generation of 5-hydroxymethylcytidine 5′-monophosphate and 5-hydroxymethylcytosine in Streptoverticillium rimofaciens ZJU5119. ChemBioChem 9, 1286–1294 (2008)
Erlanger, B. F. & Beiser, S. M. Antibodies specific for ribonucleosides and ribonucleotides and their reaction with DNA. Proc. Natl Acad. Sci. USA 52, 68–74 (1964)
de Vries, W. N. et al. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis 26, 110–112 (2000)
Ohbo, K. et al. Identification and characterization of stem cells in prepubertal spermatogenesis in mice. Dev. Biol. 258, 209–225 (2003)
Ge, Y. Z. et al. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 279, 25447–25454 (2004)
Liu, P., Jenkins, N. A. & Copeland, N. G. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13, 476–484 (2003)
Rodriguez, C. I. et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP . Nature Genet. 25, 139–140 (2000)
Lomelí, H., Ramos-Mejia, V., Gertsenstein, M., Lobe, C. G. & Nagy, A. Targeted insertion of Cre recombinase into the TNAP gene: excision in primordial germ cells. Genesis 26, 116–117 (2000)
Lewandoski, M., Wassarman, K. M. & Martin, G. R. Zp3–cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr. Biol. 7, 148–151 (1997)
Szabó, P. E., Hubner, K., Scholer, H. & Mann, J. R. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech. Dev. 115, 157–160 (2002)
Kimura, Y. & Yanagimachi, R. Intracytoplasmic sperm injection in the mouse. Biol. Reprod. 52, 709–720 (1995)
Abràmoff, M. D., Magalhães, P. J. & Ram, S. J. Image processing with ImageJ. Biophotonics Int. 11, 36–42 (2004)
Hajkova, P. et al. DNA-methylation analysis by the bisulfite-assisted genomic sequencing method. Methods Mol. Biol. 200, 143–154 (2002)
Weber, M. et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature Genet. 37, 853–862 (2005)
Tsumura, A. et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814 (2006)
Wakayama, T., Perry, A. C., Zuccotti, M., Johnson, K. R. & Yanagimachi, R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 394, 369–374 (1998)
Acknowledgements
We thank C. Walsh and M. Rots for critical reading of the manuscript, J. Walter for discussions, H. Qi for providing cDNA of mouse oocytes, R. Zhang & Q. Cui for Tet3 cDNA, L. Li for help with 5hmCMP synthesis, Shanghai Research Center for Model Organisms for blastocyst injection, and J. Gao for mouse work. This study was supported by grants from the Ministry of Science and Technology China (2007CB947503 to G.-L.X., 2007CB947101 to J.L., and 2009CB941101 to G.-L.X. and J.L.), National Science Foundation of China (30730059 to G.-L.X. and 30871430 to J.L.), the Chinese Academy of Sciences (XDA01010301 to G.-L.X.; XDA01010403 and KSCX2-YW-R-110 to J.L.) and the NIH (GM078458 to Y.G.S.).
Author information
Authors and Affiliations
Contributions
G.-L.X. and J.L. conceived the projects. Y.G.S., H.-P.W. and G.-L.X. contributed to the knockout design. F.G., T.-P.G., H.-P.W., G.-F.X., and W.L. performed the experiments on early embryos. X.H. and Z.D. contributed to the synthesis of the 5hmC hapten. H.Y. and L.S. performed the nuclear transfer and embryo transfer experiments. S.-g.J., K.I., P.E.S., G.P.P. and Z.-G.X. characterized Tet3 expression in PGCs and ovaries. G.-L.X. wrote and G.P.P. revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-16 with legends, Supplementary Tables 1-6 and additional references. (PDF 10491 kb)
Rights and permissions
About this article
Cite this article
Gu, TP., Guo, F., Yang, H. et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 (2011). https://doi.org/10.1038/nature10443
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10443
This article is cited by
-
Auto-suppression of Tet dioxygenases protects the mouse oocyte genome from oxidative demethylation
Nature Structural & Molecular Biology (2024)
-
Machine learning-based clustering to identify the combined effect of the DNA fragmentation index and conventional semen parameters on in vitro fertilization outcomes
Reproductive Biology and Endocrinology (2023)
-
Base editing-mediated one-step inactivation of the Dnmt gene family reveals critical roles of DNA methylation during mouse gastrulation
Nature Communications (2023)
-
Dynamics of DNA hydroxymethylation and methylation during mouse embryonic and germline development
Nature Genetics (2023)
-
The phosphorylation to acetylation/methylation cascade in transcriptional regulation: how kinases regulate transcriptional activities of DNA/histone-modifying enzymes
Cell & Bioscience (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.