Abstract
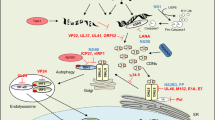
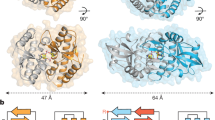
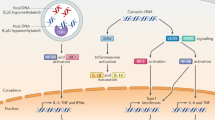
The innate immune system detects infection by using germline-encoded receptors that are specific for conserved microbial molecules. The recognition of microbial ligands leads to the production of cytokines, such as type I interferons (IFNs), that are essential for successful pathogen elimination. Cytosolic detection of pathogen-derived DNA is one major mechanism of inducing IFN production1,2, and this process requires signalling through TANK binding kinase 1 (TBK1) and its downstream transcription factor, IFN-regulatory factor 3 (IRF3). In addition, a transmembrane protein called STING (stimulator of IFN genes; also known as MITA, ERIS, MPYS and TMEM173) functions as an essential signalling adaptor, linking the cytosolic detection of DNA to the TBK1–IRF3 signalling axis3,4,5,6,7. Recently, unique nucleic acids called cyclic dinucleotides, which function as conserved signalling molecules in bacteria8, have also been shown to induce a STING-dependent type I IFN response9,10,11,12. However, a mammalian sensor of cyclic dinucleotides has not been identified. Here we report evidence that STING itself is an innate immune sensor of cyclic dinucleotides. We demonstrate that STING binds directly to radiolabelled cyclic diguanylate monophosphate (c-di-GMP), and we show that unlabelled cyclic dinucleotides, but not other nucleotides or nucleic acids, compete with c-di-GMP for binding to STING. Furthermore, we identify mutations in STING that selectively affect the response to cyclic dinucleotides without affecting the response to DNA. Thus, STING seems to function as a direct sensor of cyclic dinucleotides, in addition to its established role as a signalling adaptor in the IFN response to cytosolic DNA. Cyclic dinucleotides have shown promise as novel vaccine adjuvants and immunotherapeutics9,13, and our results provide insight into the mechanism by which cyclic dinucleotides are sensed by the innate immune system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ishii, K. J. et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nature Immunol. 7, 40–48 (2006)
Stetson, D. B. & Medzhitov, R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24, 93–103 (2006)
Ishikawa, H. & Barber, G. N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008)
Ishikawa, H., Ma, Z. & Barber, G. N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 (2009)
Jin, L. et al. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 28, 5014–5026 (2008)
Sun, W. et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl Acad. Sci. USA 106, 8653–8658 (2009)
Zhong, B. et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008)
Tamayo, R., Pratt, J. T. & Camilli, A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61, 131–148 (2007)
Karaolis, D. K. et al. Bacterial c-di-GMP is an immunostimulatory molecule. J. Immunol. 178, 2171–2181 (2007)
McWhirter, S. M. et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J. Exp. Med. 206, 1899–1911 (2009)
Sauer, J. D. et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79, 688–694 (2011)
Woodward, J. J., Iavarone, A. T. & Portnoy, D. A. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705 (2010)
Chen, W., Kuolee, R. & Yan, H. The potential of 3′,5′-cyclic diguanylic acid (c-di-GMP) as an effective vaccine adjuvant. Vaccine 28, 3080–3085 (2010)
Witte, G., Hartung, S., Buttner, K. & Hopfner, K. P. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell 30, 167–178 (2008)
Janeway, C. A., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13 (1989)
Unterholzner, L. et al. IFI16 is an innate immune sensor for intracellular DNA. Nature Immunol. 11, 997–1004 (2010)
Ablasser, A. et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nature Immunol. 10, 1065–1072 (2009)
Chiu, Y. H., Macmillan, J. B. & Chen, Z. J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138, 576–591 (2009)
Chodosh, L. A. in Current Protocols in Molecular Biology Ch. 12, 12.5.1–12.5.8 (Wiley, 2001)
Takaoka, A. et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 (2007)
Merighi, M., Lee, V. T., Hyodo, M., Hayakawa, Y. & Lory, S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65, 876–895 (2007)
Broz, P., von Moltke, J., Jones, J. W., Vance, R. E. & Monack, D. M. Differential requirement for caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe 8, 471–483 (2010)
Kulesekara, H. et al. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl Acad. Sci. USA 103, 2839–2844 (2006)
Hyodo, M. & Hayakawa, Y. An improved method for synthesizing cyclic bis(3′-5′)diguanylic acid (c-di-GMP). Bull. Chem. Soc. Jpn 77, 2089–2093 (2004)
Simm, R., Remminghorst, U., Ahmad, I., Zakikhany, K. & Romling, U. A role for the EAL-like protein STM1344 in regulation of CsgD expression and motility in Salmonella enterica serovar Typhimurium. J. Bacteriol. 191, 3928–3937 (2009)
De, N. et al. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 6, e67 (2008)
Acknowledgements
We thank H. Carlson, K. Collins, S. McWhirter, D. Raulet, K. Sjölander and members of the Vance, Barton and Portnoy laboratories at the University of California, Berkeley, for advice and discussions. We thank J. Woodward and D. Portnoy for their gift of purified c-di-AMP. Work in R.E.V.’s laboratory is supported by investigator awards from the Burroughs Wellcome Fund and the Cancer Research Institute and by National Institutes of Health (NIH) grants AI075039, AI080749 and AI063302. D.L.B. is supported by an NIH National Research Service Award fellowship F32 (AI091100).
Author information
Authors and Affiliations
Contributions
D.L.B. performed luciferase assays and quantitative RT–PCR, generated c-di-[32P]GMP, purified recombinant STING, performed c-di-[32P]GMP binding assays and transduced gt bone marrow macrophages. K.M.M. generated truncation mutations and performed luciferase assays. K.S.-T. generated point mutants and performed luciferase assays. D.L.B., K.M.M. and R.E.V. participated in study design and data analysis. D.L.B. and R.E.V. wrote the paper. B.E. contributed to protein purification methods. J.S.I. contributed to the design of the equilibrium dialysis experiments and the analysis of binding data. M.H. and Y.H. synthesized c-di-GMP.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-5 with legends and Supplementary Table 1. (PDF 1342 kb)
Rights and permissions
About this article
Cite this article
Burdette, D., Monroe, K., Sotelo-Troha, K. et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011). https://doi.org/10.1038/nature10429
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10429
This article is cited by
-
STING-dependent trained immunity contributes to host defense against Clostridium perfringens infection via mTOR signaling
Veterinary Research (2024)
-
Cytosolic DNA sensors in neurodegenerative diseases: from physiological defenders to pathological culprits
EMBO Molecular Medicine (2024)
-
Cholesterol-binding motifs in STING that control endoplasmic reticulum retention mediate anti-tumoral activity of cholesterol-lowering compounds
Nature Communications (2024)
-
ESCRT-dependent STING degradation inhibits steady-state and cGAMP-induced signalling
Nature Communications (2023)
-
Epigenetic state determines the in vivo efficacy of STING agonist therapy
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.