Abstract
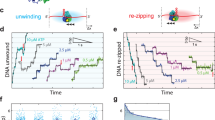
Helicases are vital enzymes that carry out strand separation of duplex nucleic acids during replication, repair and recombination1,2. Bacteriophage T7 gene product 4 is a model hexameric helicase that has been observed to use dTTP, but not ATP, to unwind double-stranded (ds)DNA as it translocates from 5′ to 3′ along single-stranded (ss)DNA2,3,4,5,6. Whether and how different subunits of the helicase coordinate their chemo-mechanical activities and DNA binding during translocation is still under debate1,7. Here we address this question using a single-molecule approach to monitor helicase unwinding. We found that T7 helicase does in fact unwind dsDNA in the presence of ATP and that the unwinding rate is even faster than that with dTTP. However, unwinding traces showed a remarkable sawtooth pattern where processive unwinding was repeatedly interrupted by sudden slippage events, ultimately preventing unwinding over a substantial distance. This behaviour was not observed with dTTP alone and was greatly reduced when ATP solution was supplemented with a small amount of dTTP. These findings presented an opportunity to use nucleotide mixtures to investigate helicase subunit coordination. We found that T7 helicase binds and hydrolyses ATP and dTTP by competitive kinetics such that the unwinding rate is dictated simply by their respective maximum rates Vmax, Michaelis constants KM and concentrations. In contrast, processivity does not follow a simple competitive behaviour and shows a cooperative dependence on nucleotide concentrations. This does not agree with an uncoordinated mechanism where each subunit functions independently, but supports a model where nearly all subunits coordinate their chemo-mechanical activities and DNA binding. Our data indicate that only one subunit at a time can accept a nucleotide while other subunits are nucleotide-ligated and thus they interact with the DNA to ensure processivity. Such subunit coordination may be general to many ring-shaped helicases and reveals a potential mechanism for regulation of DNA unwinding during replication.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
06 October 2011
The labelling of Fig. 1d and Fig. 3a was corrected.
References
Singleton, M. R., Dillingham, M. S. & Wigley, D. B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50 (2007)
Patel, S. S. & Picha, K. M. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69, 651–697 (2000)
Donmez, I. & Patel, S. S. Mechanisms of a ring shaped helicase. Nucleic Acids Res. 34, 4216–4224 (2006)
Matson, S. W., Tabor, S. & Richardson, C. C. The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J. Biol. Chem. 258, 14017–14024 (1983)
Matson, S. W. & Richardson, C. C. DNA-dependent nucleoside 5′-triphosphatase activity of the gene 4 protein of bacteriophage T7. J. Biol. Chem. 258, 14009–14016 (1983)
Hingorani, M. M. & Patel, S. S. Cooperative interactions of nucleotide ligands are linked to oligomerization and DNA binding in bacteriophage T7 gene 4 helicases. Biochemistry 35, 2218–2228 (1996)
Lyubimov, A. Y., Strycharska, M. & Berger, J. M. The nuts and bolts of ring-translocase structure and mechanism. Curr. Opin. Struct. Biol. 21, 240–248 (2011)
Lee, S. J. & Richardson, C. C. Molecular basis for recognition of nucleoside triphosphate by gene 4 helicase of bacteriophage T7. J. Biol. Chem. 285, 31462–31471 (2010)
Johnson, D. S., Bai, L., Smith, B. Y., Patel, S. S. & Wang, M. D. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell 129, 1299–1309 (2007)
Myong, S., Rasnik, I., Joo, C., Lohman, T. M. & Ha, T. Repetitive shuttling of a motor protein on DNA. Nature 437, 1321–1325 (2005)
Myong, S., Bruno, M. M., Pyle, A. M. & Ha, T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science 317, 513–516 (2007)
Sun, B. et al. Impediment of E. coli UvrD by DNA-destabilizing force reveals a strained-inchworm mechanism of DNA unwinding. EMBO J. 27, 3279–3287 (2008)
Dessinges, M. N., Lionnet, T., Xi, X. G., Bensimon, D. & Croquette, V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc. Natl Acad. Sci. USA 101, 6439–6444 (2004)
Chemla, Y. R. et al. Mechanism of force generation of a viral DNA packaging motor. Cell 122, 683–692 (2005)
Tsay, J. M., Sippy, J., Feiss, M. & Smith, D. E. The Q motif of a viral packaging motor governs its force generation and communicates ATP recognition to DNA interaction. Proc. Natl Acad. Sci. USA 106, 14355–14360 (2009)
Singleton, M. R., Sawaya, M. R., Ellenberger, T. & Wigley, D. B. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101, 589–600 (2000)
Patel, S. S., Rosenberg, A. H., Studier, F. W. & Johnson, K. A. Large scale purification and biochemical characterization of T7 primase/helicase proteins. Evidence for homodimer and heterodimer formation. J. Biol. Chem. 267, 15013–15021 (1992)
Mathews, C. K. Biochemistry of deoxyribonucleic acid-defective amber mutants of bacteriophage T4. 3. Nucleotide pools. J. Biol. Chem. 247, 7430–7438 (1972)
Hingorani, M. M. & Patel, S. S. Interactions of bacteriophage T7 DNA primase/helicase protein with single-stranded and double-stranded DNAs. Biochemistry 32, 12478–12487 (1993)
Liao, J. C., Jeong, Y. J., Kim, D. E., Patel, S. S. & Oster, G. Mechanochemistry of T7 DNA helicase. J. Mol. Biol. 350, 452–475 (2005)
Crampton, D. J., Mukherjee, S. & Richardson, C. C. DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol. Cell 21, 165–174 (2006)
Yu, X., Hingorani, M. M., Patel, S. S. & Egelman, E. H. DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nature Struct. Biol. 3, 740–743 (1996)
Enemark, E. J. & Joshua-Tor, L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442, 270–275 (2006)
Thomsen, N. D. & Berger, J. M. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell 139, 523–534 (2009)
Koch, S. J., Shundrovsky, A., Jantzen, B. C. & Wang, M. D. Probing protein-DNA interactions by unzipping a single DNA double helix. Biophys. J. 83, 1098–1105 (2002)
Peterman, E. J., Gittes, F. & Schmidt, C. F. Laser-induced heating in optical traps. Biophys. J. 84, 1308–1316 (2003)
Shundrovsky, A., Smith, C. L., Lis, J. T., Peterson, C. L. & Wang, M. D. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nature Struct. Mol. Biol. 13, 549–554 (2006)
Lohman, T. M., Tomko, E. J. & Wu, C. G. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nature Rev. Mol. Cell Biol. 9, 391–401 (2008)
Acknowledgements
We thank members of the Wang laboratory for critical reading of the manuscript. We also thank M. A. Hall for assistance with single-molecule assays, data acquisition and data analysis. We wish to acknowledge support from National Institutes of Health grants (GM059849 to M.D.W.; GM55310 to S.S.P.), National Science Foundation grant (MCB-0820293 to M.D.W.) and Cornell’s Molecular Biophysics Training Grant (T32GM008267) Traineeship (to D.S.J. and B.Y.S.).
Author information
Authors and Affiliations
Contributions
B.S., D.S.J., S.S.P. and M.D.W. designed the experiments. D.S.J. found the helicase slippage with ATP. B.S. carried out all single-molecule work and, together with B.Y.S., analysed and interpreted single-molecule data. G.P. performed all the ensemble experiments. M.P. and G.P. purified and analysed the wild-type and mutant T7 gp4 proteins. M.D.W. formulated the theoretical models. B.S., D.S.J., B.Y.S., S.S.P. and M.D.W. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-13 with legends, a Supplementary Discussion and additional references. (PDF 1587 kb)
Rights and permissions
About this article
Cite this article
Sun, B., Johnson, D., Patel, G. et al. ATP-induced helicase slippage reveals highly coordinated subunits. Nature 478, 132–135 (2011). https://doi.org/10.1038/nature10409
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10409
This article is cited by
-
Optical tweezers in single-molecule biophysics
Nature Reviews Methods Primers (2021)
-
A DNA packaging motor inchworms along one strand allowing it to adapt to alternative double-helical structures
Nature Communications (2021)
-
The mechanism of DNA unwinding by the eukaryotic replicative helicase
Nature Communications (2019)
-
Active DNA unwinding and transport by a membrane-adapted helicase nanopore
Nature Communications (2019)
-
Helicase promotes replication re-initiation from an RNA transcript
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.