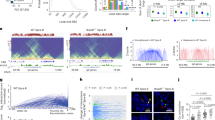
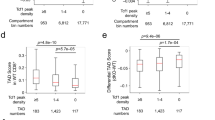
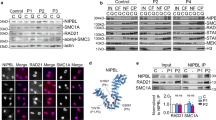
Abstract
Cohesin enables post-replicative DNA repair and chromosome segregation by holding sister chromatids together from the time of DNA replication in S phase until mitosis1. There is growing evidence that cohesin also forms long-range chromosomal cis-interactions2,3,4 and may regulate gene expression2,3,4,5,6,7,8,9,10 in association with CTCF8,9, mediator4 or tissue-specific transcription factors10. Human cohesinopathies such as Cornelia de Lange syndrome are thought to result from impaired non-canonical cohesin functions7, but a clear distinction between the cell-division-related and cell-division-independent functions of cohesion—as exemplified in Drosophila11,12,13—has not been demonstrated in vertebrate systems. To address this, here we deleted the cohesin locus Rad21 in mouse thymocytes at a time in development when these cells stop cycling and rearrange their T-cell receptor (TCR) α locus (Tcra). Rad21-deficient thymocytes had a normal lifespan and retained the ability to differentiate, albeit with reduced efficiency. Loss of Rad21 led to defective chromatin architecture at the Tcra locus, where cohesion-binding sites flank the TEA promoter and the Eα enhancer, and demarcate Tcra from interspersed Tcrd elements and neighbouring housekeeping genes. Cohesin was required for long-range promoter–enhancer interactions, Tcra transcription, H3K4me3 histone modifications that recruit the recombination machinery14,15 and Tcra rearrangement. Provision of pre-rearranged TCR transgenes largely rescued thymocyte differentiation, demonstrating that among thousands of potential target genes across the genome4,8,9,10, defective Tcra rearrangement was limiting for the differentiation of cohesin-deficient thymocytes. These findings firmly establish a cell-division-independent role for cohesin in Tcra locus rearrangement and provide a comprehensive account of the mechanisms by which cohesin enables cellular differentiation in a well-characterized mammalian system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nasmyth, K. & Haering, C. H. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558 (2009)
Hadjur, S. et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460, 410–413 (2009)
Degner, S. C. et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc. Natl Acad. Sci. USA 10.1073/pnas.1019391108 (23 May 2011)
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010)
Hagstrom, K. A. & Meyer, B. J. Condensin and cohesin: more than chromosome compactor and glue. Nature Rev. Genet. 4, 520–534 (2003)
Rollins, R. A., Morcillo, P. & Dorsett, D. Nipped-B, a Drosophila homologue of chromosomal adherins participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152, 577–593 (1999)
Strachan, T. Cornelia de Lange syndrome and the link between chromosomal function, DNA repair and developmental gene regulation. Curr. Opin. Genet. Dev. 15, 258–264 (2005)
Parelho, V. et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433 (2008)
Wendt, K. S. et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801 (2008)
Schmidt, D. et al. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 20, 578–588 (2010)
Schuldiner, O. et al. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev. Cell 14, 227–238 (2008)
Pauli, A. et al. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev. Cell 14, 239–251 (2008)
Pauli, A. et al. A direct role for cohesin in gene regulation and ecdysone response in Drosophila salivary glands. Curr. Biol. 20, 1787–1798 (2010)
Matthews, A. G. et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450, 1106–1110 (2007)
Liu, Y., Subrahmanyam, R., Chakraborty, T., Sen, R. & Desiderio, S. A plant homeodomain in Rag-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity 27, 561–571 (2007)
Kisielow, P. & von Boehmer, H. Development and selection of T cells: facts and puzzles. Adv. Immunol. 58, 87–209 (1995)
Stanhope-Baker, P., Hudson, K. M., Shaffer, A. L., Constantinescu, A. & Schlissel, M. S. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro . Cell 85, 887–897 (1996)
Krangel, M. S. Mechanics of T cell receptor gene rearrangement. Curr. Opin. Immunol. 21, 133–139 (2009)
Jhunjhunwala, S., van Zelm, M. C., Peak, M. M. & Murre, C. Chromatin architecture and the generation of antigen receptor diversity. Cell 138, 435–448 (2009)
Desiderio, S., Lin, W. C. & Li, Z. The cell cycle and V(D)J recombination. Curr. Top. Microbiol. Immunol. 217, 45–59 (1996)
Brekelmans, P. et al. Transferrin receptor expression as a marker of immature cycling thymocytes in the mouse. Cell. Immunol. 159, 331–339 (1994)
Huesmann, M., Scott, B., Kisielow, P. & von Boehmer, H. Kinetics and efficacy of positive selection in the thymus of normal and T cell receptor transgenic mice. Cell 66, 533–540 (1991)
Lee, P. P. et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001)
Diaz, P., Cado, D. & Winoto, A. A locus control region in the T cell receptor α/δ locus. Immunity 1, 207–217 (1994)
Magdinier, F., Yusufzai, T. M. & Felsenfeld, G. Both CTCF-dependent and -independent insulators are found between the mouse T cell receptor α and Dad1 genes. J. Biol. Chem. 279, 25381–25389 (2004)
Zhong, X. P. & Krangel, M. S. Enhancer-blocking activity within the DNAse I hypersensitivity site 2 to 6 region between the T cell receptor α and Dad1 genes. J. Immunol. 163, 295–300 (1999)
Abarrategui, I. & Krangel, M. S. Germline transcription: a key regulator of accessibility and recombination. Adv. Exp. Med. Biol. 650, 93–102 (2009)
Ji, Y. et al. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell 141, 419–431 (2010)
Guo, J. et al. Regulation of the TCRα repertoire by the survival window of CD4+CD8+ thymocytes. Nature Immunol. 3, 469–476 (2002)
Dekker, J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nature Methods 3, 17–21 (2006)
Spanopoulou, E. et al. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 8, 1030–1042 (1994)
Hogquist, K. A. et al. T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994)
Kaye, J. et al. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature 341, 746–749 (1989)
Abarrategui, I. & Krangel, M. S. Regulation of T cell receptor-α gene recombination by transcription. Nature Immunol. 7, 1109–1115 (2006)
Jackson, A., Kondilis, H. D., Khor, B., Sleckman, B. P. & Krangel, M. S. Regulation of T cell receptor β allelic exclusion at a level beyond accessibility. Nature Immunol. 6, 189–197 (2005)
Toyoda, Y. & Yanagida, M. Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol. Biol. Cell 17, 2287–2302 (2006)
Kibbe, W. A. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 35, W43–W46 (2007)
Fejes, A. P. et al. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics 24, 1729–1730 (2008)
Lee, Y. N. et al. Differential utilization of T cell receptor TCRα/TCRδ locus variable region gene segments is mediated by accessibility. Proc. Natl Acad. Sci. USA 106, 17487–17492 (2009)
Acknowledgements
We thank S. Hadjur, D. Tough, L. Williams, Z. Webster, J. Godwin and H.-Y. Shih for help and advice, L. Game and M. Jones for high-throughput sequencing, A. Giess for sequence alignment, and J. Elliott and P. Hexley for cell sorting. Supported by the Medical Research Council, UK (V.S., T.L., H.M.-B., K.E.B., T.C., A.T., L.A., A.G.F., K.N., M.M.), the European Union FP6 integrated project HEROIC (H.M.), EU and the Marie Curie Research Training Network Chromatin Plasticity (H.M.-B.), the Boehringer Ingelheim Fonds (T.L.), the Wellcome Trust (D.J.A., K.N.) and the National Institutes of Health (B.H., M.S.K., G.T., D.G.S.). D.G.S. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
V.S. and M.M. conceived the study with critical input from D.G.S., L.A., A.G.F., M.S.K. and K.N., V.S., B.H., K.T.-K., T.L., H.M.-B., K.E.B., G.T., K.H. and M.M. conducted experiments, K.T.-K., D.J.A., K.N., G.T. and D.G.S. designed and generated novel materials, T.C., A.T. and H.M. analysed data, V.S. and M.M. wrote the paper and all authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-8 with legends and additional references. (PDF 5104 kb)
Rights and permissions
About this article
Cite this article
Seitan, V., Hao, B., Tachibana-Konwalski, K. et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 476, 467–471 (2011). https://doi.org/10.1038/nature10312
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10312
This article is cited by
-
Boundary stacking interactions enable cross-TAD enhancer–promoter communication during limb development
Nature Genetics (2024)
-
Genome control by SMC complexes
Nature Reviews Molecular Cell Biology (2023)
-
Super-resolution visualization of chromatin loop folding in human lymphoblastoid cells using interferometric photoactivated localization microscopy
Scientific Reports (2022)
-
CTCF orchestrates long-range cohesin-driven V(D)J recombinational scanning
Nature (2020)
-
Spt5-mediated enhancer transcription directly couples enhancer activation with physical promoter interaction
Nature Genetics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.