Abstract
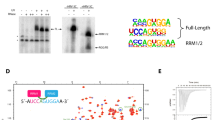
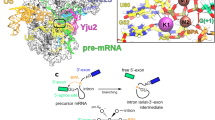
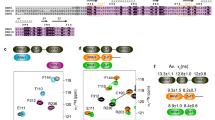
Many cellular functions involve multi-domain proteins, which are composed of structurally independent modules connected by flexible linkers. Although it is often well understood how a given domain recognizes a cognate oligonucleotide or peptide motif, the dynamic interaction of multiple domains in the recognition of these ligands remains to be characterized. Here we have studied the molecular mechanisms of the recognition of the 3′-splice-site-associated polypyrimidine tract RNA by the large subunit of the human U2 snRNP auxiliary factor (U2AF65)1,2,3 as a key early step in pre-mRNA splicing4. We show that the tandem RNA recognition motif domains of U2AF65 adopt two remarkably distinct domain arrangements in the absence or presence of a strong (that is, high affinity) polypyrimidine tract. Recognition of sequence variations in the polypyrimidine tract RNA involves a population shift between these closed and open conformations. The equilibrium between the two conformations functions as a molecular rheostat that quantitatively correlates the natural variations in polypyrimidine tract nucleotide composition, length and functional strength to the efficiency to recruit U2 snRNP to the intron during spliceosome assembly1,5,6,7,8. Mutations that shift the conformational equilibrium without directly affecting RNA binding modulate splicing activity accordingly. Similar mechanisms of cooperative multi-domain conformational selection may operate more generally in the recognition of degenerate nucleotide or amino acid motifs by multi-domain proteins9,10.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
The coordinates of the open RNA-bound conformation of RRM1–RRM2 and the closed conformation in the absence of RNA are deposited in the Protein Data Bank with accession codes 2YH1 and 2YH0, respectively. All structural ensembles with explicit spin labels are available from the authors upon request.
References
Zamore, P. D., Patton, J. G. & Green, M. R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355, 609–614 (1992)
Banerjee, H., Rahn, A., Davis, W. & Singh, R. Sex lethal and U2 small nuclear ribonucleoprotein auxiliary factor (U2AF65) recognize polypyrimidine tracts using multiple modes of binding. RNA 9, 88–99 (2003)
Banerjee, H. et al. The conserved RNA recognition motif 3 of U2 snRNA auxiliary factor (U2AF65) is essential in vivo but dispensible for activity in vitro . RNA 10, 240–253 (2004)
Wahl, M. C., Will, C. L. & Lührmann, R. The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718 (2009)
Reed, R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 3, 2113–2123 (1989)
Roscigno, R. F., Weiner, M. & Garcia-Blanco, M. A. A mutational analysis of the polypyrimidine tract of introns. Effects of sequence differences in pyrimidine tracts on splicing. J. Biol. Chem. 268, 11222–11229 (1993)
Singh, R., Valcárcel, J. & Green, M. R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268, 1173–1176 (1995)
Coolidge, C. J., Seely, R. J. & Patton, J. G. Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res. 25, 888–896 (1997)
Taverna, S. D., Li, H., Ruthenburg, A. J., Allis, C. D. & Patel, D. J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature Struct. Mol. Biol. 14, 1025–1040 (2007)
Seet, B. T., Dikic, I., Zhou, M. M. & Pawson, T. Reading protein modifications with interaction domains. Nature Rev. Mol. Cell Biol. 7, 473–483 (2006)
Ito, T., Muto, Y., Green, M. R. & Yokoyama, S. Solution structures of the first and second RNA-binding domains of human U2 small nuclear ribonucleoprotein particle auxiliary factor (U2AF(65)). EMBO J. 18, 4523–4534 (1999)
Sickmier, E. A. et al. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol. Cell 23, 49–59 (2006)
Simon, B., Madl, T., Mackereth, C. D., Nilges, M. & Sattler, M. An efficient protocol for NMR-spectroscopy-based structure determination of protein complexes in solution. Angew. Chem. Int. Edn Engl. 49, 1967–1970 (2010)
Boehr, D. D., Nussinov, R. & Wright, P. E. The role of dynamic conformational ensembles in biomolecular recognition. Nature Chem. Biol. 5, 789–796 (2009)
Zhang, Q., Stelzer, A. C., Fisher, C. K. & Al-Hashimi, H. M. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature 450, 1263–1267 (2007)
Lange, O. F. et al. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science 320, 1471–1475 (2008)
Li, P., Martins, I. R., Amarasinghe, G. K. & Rosen, M. K. Internal dynamics control activation and activity of the autoinhibited Vav DH domain. Nature Struct. Mol. Biol. 15, 613–618 (2008)
Korzhnev, D. M., Religa, T. L., Banachewicz, W., Fercht, A. R. & Kay, L. E. A transient and low-populated protein-folding intermediate at atomic resolution. Science 329, 1312–1316 (2010)
Henzler-Wildman, K. A. et al. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature 450, 913–916 (2007)
Shoemaker, B. A., Portman, J. J. & Wolynes, P. G. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl Acad. Sci. USA 97, 8868–8873 (2000)
Hammes, G. G., Chang, Y.-C. & Oas, T. G. Conformational selection or induced fit: a flux description of reaction mechanism. Proc. Natl Acad. Sci. USA 106, 13737–13741 (2009)
Webby, C. J. et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93 (2009)
Soares, L. M., Zanier, K., Mackereth, C., Sattler, M. & Valcárcel, J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science 312, 1961–1965 (2006)
Rückert, M. & Otting, G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 122, 7793–7797 (2000)
Guth, S., Tange, T., Ø, Kellenberger, E. & Valcárcel, J. Dual function for U2AF(35) in AG-dependent pre-mRNA splicing. Mol. Cell. Biol. 21, 7673–7681 (2001)
Valcárcel, J., Martínez, C. & Green, M. R. Functional analysis of splicing factors and regulators. In mRNA Formation and Function 31–53 (Elsevier, 1997)
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995)
Johnson, B. A. & Blevins, R. A. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994)
Farrow, N. A. et al. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 (1994)
Yang, D., Venters, R. A., Mueller, G. A., Choy, W. Y. & Kay, L. E. TROSY-based HNCO pulse sequences for the measurement of 1HN-15N, 15N-13CO, 1HN-13CO, 13CO-13Cα and 1HN-13Cα dipolar couplings in 15N, 13C, 2H-labelled proteins. J. Biomol. NMR 14, 333–343 (1999)
Iwahara, J., Schwieters, C. D. & Clore, G. M. Ensemble approach for NMR structure refinement against 1H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J. Am. Chem. Soc. 126, 5879–5896 (2004)
Ramos, A. & Varani, G. A new method to detect long-range protein-RNA contacts: NMR detection of electron-proton relaxation induced by nitroxide spin-labeled RNA. J. Am. Chem. Soc. 120, 10992–10993 (1998)
Nilges, M. Calculation of protein structures with ambiguous distance restraints. Automated assignment of ambiguous NOE crosspeaks and disulphide connectivities. J. Mol. Biol. 245, 645–660 (1995)
Brünger, A. T. et al. Crystallography 1 NMR system : A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Cornilescu, G., Delaglio, F. & Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 (1999)
Buckle, P. E. et al. The resonant mirror: a novel optical sensor for direct sensing of biomolecular interactions part II: applications. Biosens. Bioelectron. 8, 355–363 (1993)
Acknowledgements
We thank F. Gabel, M. Nilges, C. Griesinger, J. Müller and K. Scheffzek for discussions, and H. Tilgner for analysis of natural Py tract sequences. C.D.M. acknowledges support by EMBO Long Term Fellowship, ICSN and Aquitaine regional government. T.M. thanks the Austrian Science Fund (FWF) and EMBO for postdoctoral fellowships. We thank the EU NMR LSF in Frankfurt and the Bavarian NMR Centre (BNMRZ) in Munich for NMR measurement time. This work was supported by the European Commission, grants 3D Repertoire, FSG-V-RNA and NIM3 No. 226507 (M.S.), EURASNET, AICR and Fundación Marcelino Botín (J.V.).
Author information
Authors and Affiliations
Contributions
C.D.M., S.B., K.Z. and A.G. cloned and purified native and nitroxyl-labelled proteins. C.D.M., K.Z., B.S. and T.M. collected, processed and analysed NMR spectroscopy data. C.D.M., B.S. and T.M. calculated and analysed structural ensembles. S.B. performed in vitro splicing assays. V.R. performed ITC. J.V. and M.S. contributed to study design. C.D.M. and M.S. wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Text, Supplementary References, Supplementary Tables 1-3 and Supplementary Figures 1-16 with legends. (PDF 21889 kb)
Rights and permissions
About this article
Cite this article
Mackereth, C., Madl, T., Bonnal, S. et al. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature 475, 408–411 (2011). https://doi.org/10.1038/nature10171
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10171
This article is cited by
-
Structural basis for specific RNA recognition by the alternative splicing factor RBM5
Nature Communications (2023)
-
Reliability and accuracy of single-molecule FRET studies for characterization of structural dynamics and distances in proteins
Nature Methods (2023)
-
Nucleotide-amino acid π-stacking interactions initiate photo cross-linking in RNA-protein complexes
Nature Communications (2022)
-
SPF45/RBM17-dependent, but not U2AF-dependent, splicing in a distinct subset of human short introns
Nature Communications (2021)
-
Mechanism and evolution of the Zn-fingernail required for interaction of VARP with VPS29
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.