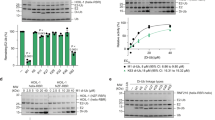
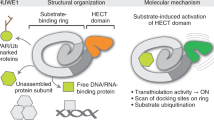
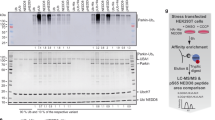
Abstract
Although the functional interaction between ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s) is essential in ubiquitin (Ub) signalling, the criteria that define an active E2–E3 pair are not well established. The human E2 UBCH7 (also known as UBE2L3) shows broad specificity for HECT-type E3s1, but often fails to function with RING E3s in vitro despite forming specific complexes2,3,4. Structural comparisons of inactive UBCH7–RING complexes with active UBCH5–RING complexes reveal no defining differences3,4, highlighting a gap in our understanding of Ub transfer. Here we show that, unlike many E2s that transfer Ub with RINGs, UBCH7 lacks intrinsic, E3-independent reactivity with lysine, explaining its preference for HECTs. Despite lacking lysine reactivity, UBCH7 exhibits activity with the RING-in-between-RING (RBR) family of E3s that includes parkin (also known as PARK2) and human homologue of ariadne (HHARI; also known as ARIH1)5,6. Found in all eukaryotes7, RBRs regulate processes such as translation8 and immune signalling9. RBRs contain a canonical C3HC4-type RING, followed by two conserved Cys/His-rich Zn2+-binding domains, in-between-RING (IBR) and RING2 domains, which together define this E3 family7. We show that RBRs function like RING/HECT hybrids: they bind E2s via a RING domain, but transfer Ub through an obligate thioester-linked Ub (denoted ∼Ub), requiring a conserved cysteine residue in RING2. Our results define the functional cadre of E3s for UBCH7, an E2 involved in cell proliferation10 and immune function11, and indicate a novel mechanism for an entire class of E3s.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Anan, T. et al. Human ubiqutin-protein ligase Nedd4: expression, subcellular localization and selective interaction with ubiquitin-conjugating enzymes. Genes Cells 3, 751–763 (1998)
Zheng, N., Wang, P., Jeffrey, P. D. & Pavletich, N. P. Structure of a c-Cbl–UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533–539 (2000)
Brzovic, P. S. et al. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl Acad. Sci. USA 100, 5646–5651 (2003)
Huang, A. et al. E2–c-Cbl recognition is necessary but not sufficent for ubiquitination activity. J. Mol. Biol. 385, 507–519 (2009)
Capili, A. D., Edghill, E. L., Wu, K. & Borden, K. Structure of the C-terminal RING finger from a RING-IBR-RING/TRIAD motif reveals a novel zinc-binding domain distinct from a RING. J. Mol. Biol. 340, 1117–1129 (2004)
Shimura, H. et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nature Genet. 25, 302–305 (2000)
Marin, I., Lucas, J. I., Gradilla, A. C. & Ferrus, A. Parkin and relatives: the RBR family of ubiquitin ligases. Physiol. Genomics 17, 253–263 (2004)
Tan, N. G. et al. Human homologue of ariadne promotes the ubiquitylation of translation initiation factor 4E homologous protein, 4EHP. FEBS Lett. 554, 501–504 (2003)
Tokunaga, F. et al. Involvement of linear polyubuitylation of NEMO in NF-κB activation. Nature Cell Biol. 11, 123–132 (2009)
Whitcomb, E. A., Dudek, E. J., Liu, Q. & Taylor, A. Novel control of S phase of the cell cycle by ubiquitin-conjugating enzyme H7. Mol. Biol. Cell 20, 1–9 (2009)
Fransen, K. et al. Analysis of SNPs with an effect on gene expression identifies UBE2L3 and BCL3 as potential new risk genes for Crohn’s disease. Hum. Mol. Genet. 19, 3482–3488 (2010)
Pickart, C. M. & Rose, I. A. Functional heterogeneity of ubiquitin carrier proteins. J. Biol. Chem. 260, 1573–1581 (1985)
Wang, X. et al. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-1 by viral E3 ligase mK3. J. Cell Biol. 177, 613–624 (2007)
Cadwell, K. & Coscoy, L. Ubiquitination of nonlysine residues by a viral E3 ubiquitin ligase. Science 309, 127–130 (2005)
Christensen, D. E., Brzovic, P. S. & Klevit, R. E. E2–BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nature Struct. Mol. Biol. 14, 941–948 (2007)
Rodrigo-Brenni, M. C. & Morgan, D. O. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell 130, 127–139 (2007)
Jones, D., Crowe, E., Stevens, T. A. & Candido, E. P. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes and ubiquitin-like proteins. Genome Biology 3, RESEARCH0002 (2002)
Yunus, A. A. & Lima, C. D. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nature Struct. Mol. Biol. 13, 491–499 (2006)
Wu, P. Y. et al. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 22, 5241–5250 (2003)
Sakata, E. et al. Crystal structure of the UbcH5b∼ubiquitin intermediate: insight into the formation of the self-assembled E2∼Ub conjugates. Structure 18, 138–147 (2010)
Ozkan, E., Yu, H. & Deisenhofer, J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc. Natl Acad. Sci. USA 102, 18890–18895 (2005)
Kamadurai, H. B. et al. Insights into ubiquitin transfer cascades from a structure of UbcH5b∼Ubiquitin-HECTNEDD4L complex. Mol. Cell 36, 1095–1102 (2009)
Ardley, H. C., Tan, N. G., Rose, S. A., Markhan, A. F. & Robinson, P. A. Features of the parkin/ariadne-like ubiquitin ligase, HHARI, that regulates its interaction with the ubiquitin-conjugating enzyme Ubch7. J. Biol. Chem. 276, 19640–19647 (2001)
Eddins, M. J., Carlile, C. M., Gomez, K. M., Pickart, C. M. & Wolberger, C. Mms2–Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nature Struct. Mol. Biol. 13, 915–920 (2006)
Fallon, L. et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K–Akt signalling. Nature Cell Biol. 8, 834–842 (2006)
Joch, M. et al. Parkin-mediated monoubiquitination of the PDZ protein PICK1 regulates the activity of acid-sensing ion channels. Mol. Biol. Cell 18, 3105–3118 (2007)
Chen, D. et al. Parkin mono-ubiquitinates Bcl-2 and regulates autophagy. J. Biol. Chem. 285, 38214–38223 (2010)
Maruyama, M. et al. Novel mutations, pseudo-dominant inheritance, and possible familial affects in patients with autosomal recessive juvenille parkinsonism. Ann. Neurol. 48, 245–250 (2000)
Diao, J., Zhang, Y., Huibregtse, J. M., Zhou, D. & Chen, J. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nature Struct. Mol. Biol. 15, 65–70 (2008)
Dastur, A., Beaudenon, S., Kelley, M., Krug, R. M. & Huibregtse, M. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J. Biol. Chem. 281, 4334–4338 (2005)
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994)
Huang, L. et al. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science 286, 1321–1326 (1999)
Sheffield, P., Garrard, S. & Derewenda, Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15, 34–39 (1999)
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995)
Johnson, B. A. & Blevins, R. A. NMR View: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994)
Acknowledgements
We thank K. Stoll, M. Schwartz and N. Zheng for critical reading of the manuscript and discussions and V. Vittal, K. Stoll and P. Littlefield for technical help. We thank K. Borden, E. Fon, B. Schulman, L. Boyd, J. Chen and A. Merz for expression vectors for HHARI, parkin, E6-AP, Ubc18, human E1 and GST pull-down reagents, respectively. We acknowledge support from the National Institute of General Medical Sciences in the form of 5R01 GM088055 (R.E.K.) and T32 GM07270 (D.M.W.), and the National Science Foundation in the form of MCB0615632 (R.E.K.)
Author information
Authors and Affiliations
Contributions
D.M.W. performed all experiments. P.S.B. and A.L. contributed to the experimental design for Fig. 2 and Supplementary Fig. 5. D.M.W. and R.E.K. designed the overall study and wrote the manuscript with P.S.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-15 with legends and a Supplementary Table. (PDF 2401 kb)
Rights and permissions
About this article
Cite this article
Wenzel, D., Lissounov, A., Brzovic, P. et al. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 (2011). https://doi.org/10.1038/nature09966
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09966
This article is cited by
-
UBE2A and UBE2B are recruited by an atypical E3 ligase module in UBR4
Nature Structural & Molecular Biology (2024)
-
IL-1β turnover by the UBE2L3 ubiquitin conjugating enzyme and HECT E3 ligases limits inflammation
Nature Communications (2023)
-
Structural snapshots along K48-linked ubiquitin chain formation by the HECT E3 UBR5
Nature Chemical Biology (2023)
-
Targeting E3 ubiquitin ligases and their adaptors as a therapeutic strategy for metabolic diseases
Experimental & Molecular Medicine (2023)
-
Targeting Mitochondria as a Therapeutic Approach for Parkinson’s Disease
Cellular and Molecular Neurobiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.