Abstract
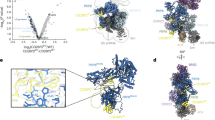
The spliceosome is a dynamic macromolecular machine that assembles on pre-messenger RNA substrates and catalyses the excision of non-coding intervening sequences (introns)1,2,3. Four of the five major components of the spliceosome, U1, U2, U4 and U5 small nuclear ribonucleoproteins (snRNPs), contain seven Sm proteins (SmB/B′, SmD1, SmD2, SmD3, SmE, SmF and SmG) in common4,5. Following export of the U1, U2, U4 and U5 snRNAs to the cytoplasm6,7, the seven Sm proteins, chaperoned by the survival of motor neurons (SMN) complex, assemble around a single-stranded, U-rich sequence called the Sm site in each small nuclear RNA (snRNA), to form the core domain of the respective snRNP particle8,9. Core domain formation is a prerequisite for re-import into the nucleus10, where these snRNPs mature via addition of their particle-specific proteins. Here we present a crystal structure of the U4 snRNP core domain at 3.6 Å resolution, detailing how the Sm site heptad (AUUUUUG) binds inside the central hole of the heptameric ring of Sm proteins, interacting one-to-one with SmE–SmG–SmD3–SmB–SmD1–SmD2–SmF. An irregular backbone conformation of the Sm site sequence combined with the asymmetric structure of the heteromeric protein ring allows each base to interact in a distinct manner with four key residues at equivalent positions in the L3 and L5 loops of the Sm fold. A comparison of this structure with the U1 snRNP at 5.5 Å resolution11,12 reveals snRNA-dependent structural changes outside the Sm fold, which may facilitate the binding of particle-specific proteins that are crucial to biogenesis of spliceosomal snRNPs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Burge, C. B., Tuschl, T. & Sharp, P. A. in The RNA World 2nd edn (eds Gesteland, R. R., Cech, T. R. & Atkins, J. F. ) 525–560 (Cold Spring Harbor Laboratory Press, 1999)
Will, C. L. & Lührmann, R. in The RNA World 3rd edn (eds Gesteland, R. F., Cech, T. R. & Atkins, J. F. ) 369–400 (Cold Spring Harbor Laboratory Press, 2006)
Yu, Y.-T., Scharl, E. C., Smith, C. M. & Steitz, J. A. in The RNA World 2nd edn (eds Gesteland, R. R.,, Cech, T. R. & Atkins, J. F. ) 487–524 (Cold Spring Harbor Laboratory Press, 1999)
Hinterberger, M., Pettersson, I. & Steitz, J. A. Isolation of small nuclear ribonucleoproteins containing U1, U2, U4, U5, and U6 RNAs. J. Biol. Chem. 258, 2604–2613 (1983)
Bringmann, P. & Lührmann, R. Purification of the individual snRNPs U1, U2, U5 and U4/U6 from HeLa cells and characterization of their protein constituents. EMBO J. 5, 3509–3516 (1986)
Mattaj, I. W. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell 46, 905–911 (1986)
Ohno, M., Segref, A., Bachi, A., Wilm, M. & Mattaj, I. W. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101, 187–198 (2000)
Meister, G., Eggert, C. & Fischer, U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 12, 472–478 (2002)
Pellizzoni, L., Yong, J. & Dreyfuss, G. Essential role for the SMN complex in the specificity of snRNP assembly. Science 298, 1775–1779 (2002)
Fischer, U., Sumpter, V., Sekine, M., Satoh, T. & Lührmann, R. Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J. 12, 573–583 (1993)
Pomeranz Krummel, D. A., Oubridge, C., Leung, A. K. W., Li, J. & Nagai, K. Crystal structure of the human spliceosomal U1 snRNP at 5.5 Å resolution. Nature 458, 475–480 (2009)
Oubridge, C., Pomeranz Krummel, D. A., Leung, A. K. W., Li, J. & Nagai, K. Interpreting a low resolution map of human U1 snRNP using anomalous scatterers. Structure 17, 930–938 (2009)
Hermann, H. et al. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein–protein interactions. EMBO J. 14, 2076–2088 (1995)
Seraphin, B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 14, 2089–2098 (1995)
Cooper, M., Johnston, L. H. & Beggs, J. D. Identification and characterization of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 14, 2066–2075 (1995)
Kambach, C. et al. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96, 375–387 (1999)
Kastner, B. & Lührmann, R. Electron microscopy of U1 small nuclear ribonucleoprotein particles: shape of the particle and position of the 5′ RNA terminus. EMBO J. 8, 277–286 (1989)
Kastner, B., Bach, M. & Lührmann, R. Electron microscopy of small nuclear ribonucleoprotein (snRNP) particles U2 and U5: evidence for a common structure-determining principle in the major U snRNP family. Proc. Natl Acad. Sci. USA 87, 1710–1714 (1990)
Törö, I. et al. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 20, 2293–2303 (2001)
Thore, S., Mayer, C., Sauter, C., Weeks, S. & Suck, D. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in Archaea and Eukarya. J. Biol. Chem. 278, 1239–1247 (2003)
Weber, G., Trowitzsch, S., Kastner, B., Lührmann, R. & Wahl, M. C. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 29, 4172–4184 (2010)
Leung, A. K. W. et al. Use of RNA tertiary interaction modules for the crystallization of the spliceosomal snRNP core domain. J. Mol. Biol. 402, 154–164 (2010)
Raker, V. A., Hartmuth, K., Kastner, B. & Lührmann, R. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol. 19, 6554–6565 (1999)
McConnell, T. S., Lokken, R. P. & Steitz, J. A. Assembly of the U1 snRNP involves interactions with the backbone of the terminal stem of U1 snRNA. RNA 9, 193–201 (2003)
Hartmuth, K., Raker, V. A., Huber, J., Branlant, C. & Lührmann, R. An unusual chemical reactivity of Sm site adenosines strongly correlates with proper assembly of core U snRNP particles. J. Mol. Biol. 285, 133–147 (1999)
Urlaub, H., Raker, V. A., Kostka, S. & Lührmann, R. Sm protein–Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 20, 187–196 (2001)
Gardner, P. P. et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 37, D137–D140 (2009)
Guthrie, C. & Patterson, B. Spliceosomal snRNAs. RNA 9, 193–201 (2003)
Draper, D. E. RNA folding: thermodynamic and molecular descriptions of the roles of ions. Biophys. J. 95, 5489–5495 (2008)
Nelissen, R. L., Will, C. L., van Venrooij, W. J. & Lührmann, R. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 13, 4113–4125 (1994)
Price, S. R., Ito, N., Oubridge, C., Avis, J. M. & Nagai, K. Crystallization of RNA–protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J. Mol. Biol. 249, 398–408 (1995)
Sauter, C. et al. Additives for the crystallization of proteins and nucleic acids. J. Cryst. Growth 196, 365–376 (1999)
Doublié, S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 (1997)
Leslie, A. G. W. The integration of macromolecular diffraction data. Acta Crystallogr. D 62, 48–57 (2006)
Evans, P. R. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006)
French, G. S. & Wilson, K. S. On the treatment of negative intensity observations. Acta Crystallogr. A 34, 517–525 (1978)
Kabsch, W. XDS. Acta Crystallogr. D 66 125–132 (2010) CrossRef
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis . Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006)
Hendrickson, W. A. & Ogata, C. M. Phase determination from multiwavelength anomalous diffraction measurements. Methods Enzymol. 276, 494–523 (1997)
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D 58, 1772–1779 (2002)
de La Fortelle, E. & Bricogne, G. Maximum likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 (1997)
Terwilliger, T. C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 (2003)
Terwilliger, T. C. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 11, 49–52 (2004)
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30, 1022–1025 (1997)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Navaza, J. Implementation of molecular replacement in AmoRe. Acta Crystallogr. D 57, 1367–1372 (2001)
Cowtan, K. D., Zhang, K. Y. J. & Main, P. In International Tables for Crystallography, Volume F. Crystallography of Biological Macromolecules (eds. Rossmann, M. G. & Arnold, E. ) 705–710 (Kluwer Academic Publishers, 2001)
Törö, I., Basquin, J., Teo-Dreher, H. & Suck, D. Archaeal Sm proteins form heptameric and hexameric complexes: crystal structures of the Sm1 and Sm2 proteins from the hyperthermophile Archaeoglobus fulgidus . J. Mol. Biol. 320, 129–142 (2002)
Collins, B. M. et al. Homomeric ring assemblies of eukaryotic Sm proteins have affinity for both RNA and DNA. Crystal structure of an oligomeric complex of yeast SmF. J. Biol. Chem. 278, 17291–17298 (2003)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot . Acta Crystallogr. D 66, 486–501 (2010)
Brunger, A. T. Version 1.2 of the Crystallography and NMR system. Nature Protocols 2, 2728–2733 (2007)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Vagin, A. et al. Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D 60, 2184–2195 (2004)
Diamond, R. On the multiple simultaneous superposition of molecular structures by rigid body transformations. Protein Sci. 1, 1279–1287 (1992)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
DeLano, W. L. The PyMOL Molecular Graphics System 〈http://www.pymol.org〉 (2002)
Acknowledgements
This work was supported by the Medical Research Council of the UK and a HFSP grant. A.K.W.L. was supported by the Overseas Research Students Awards Scheme, Canada-Cambridge Commonwealth studentship, a postgraduate scholarship from NSERC and a Junior Research Fellowship from Sidney Sussex College, Cambridge University. We thank the European Synchrotron Radiation Facility and Daresbury beamline staff for their support. We thank M. Jinek, M. Kampmann and Y. Kondo for their help with crystallization. We also thank C. Kambach, J. Avis, R. Young, S. Walke and H. Teo for laying the foundation of this project, C. Oubridge and D. Pomeranz Krummel for sharing Sm proteins and providing help and advice throughout the project, and P. Zwart for advice on twinning.
Author information
Authors and Affiliations
Contributions
A.K.W.L. and K.N. designed the constructs. A.K.W.L. crystallized the core domain, collected data and solved the structure in P6122. J.L. identified twinning and refined the structure in P31. All three authors wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-3 and Supplementary Figures 1-11 with legends. (PDF 1769 kb)
Rights and permissions
About this article
Cite this article
Leung, A., Nagai, K. & Li, J. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature 473, 536–539 (2011). https://doi.org/10.1038/nature09956
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09956
This article is cited by
-
The SMN complex drives structural changes in human snRNAs to enable snRNP assembly
Nature Communications (2023)
-
psiCLIP reveals dynamic RNA binding by DEAH-box helicases before and after exon ligation
Nature Communications (2021)
-
Structure of a pre-catalytic spliceosome
Nature (2017)
-
Cryo-electron microscopy snapshots of the spliceosome: structural insights into a dynamic ribonucleoprotein machine
Nature Structural & Molecular Biology (2017)
-
Mechanistic insights into precursor messenger RNA splicing by the spliceosome
Nature Reviews Molecular Cell Biology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.