Abstract
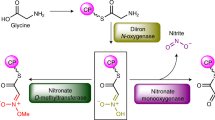
Pyrrolysine, the twenty-second amino acid found to be encoded in the natural genetic code1,2,3,4, is necessary for all of the known pathways by which methane is formed from methylamines5,6. Pyrrolysine comprises a methylated pyrroline carboxylate in amide linkage to the ε-amino group of l-lysine2,7,8. In certain Archaea, three methyltransferases initiate methanogenesis from the various methylamines9,10,11, and these enzymes are encoded by genes with an in-frame amber codon12,13 that is translated as pyrrolysine2,7,8. Escherichia coli that has been transformed with the pylTSBCD genes from methanogenic Archaea can incorporate endogenously biosynthesized pyrrolysine into proteins14. The decoding of UAG as pyrrolysine requires pylT1,6, which produces tRNAPyl (also called tRNACUA), and pylS1, which encodes a pyrrolysyl-tRNA synthetase4,15,16. The pylB, pylC and pylD genes1 are each required for tRNA-independent pyrrolysine synthesis14. Pyrrolysine is the last remaining genetically encoded amino acid with an unknown biosynthetic pathway. Here we provide genetic and mass spectrometric evidence for a pylBCD-dependent pathway in which pyrrolysine arises from two lysines. We show that a newly uncovered UAG-encoded amino acid, desmethylpyrrolysine, is made from lysine and exogenous d-ornithine in a pylC-dependent process followed by a pylD-dependent process, but it is not further converted to pyrrolysine. These results indicate that the radical S-adenosyl-l-methionine (SAM) protein PylB mediates a lysine mutase reaction that produces 3-methylornithine, which is then ligated to a second molecule of lysine by PylC before oxidation by PylD results in pyrrolysine. The discovery of lysine as the sole precursor to pyrrolysine will further inform discussions of the evolution of the genetic code and amino acid biosynthetic pathways. Furthermore, intermediates of the pathway may provide new avenues by which the pyl system can be exploited to produce recombinant proteins with useful modified residues.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Srinivasan, G., James, C. M. & Krzycki, J. A. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science 296, 1459–1462 (2002)
Hao, B. et al. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science 296, 1462–1466 (2002)
Atkins, J. F. & Gesteland, R. The 22nd amino acid. Science 296, 1409–1410 (2002)
Blight, S. K. et al. Direct charging of tRNACUA with pyrrolysine in vitro and in vivo. Nature 431, 333–335 (2004)
Krzycki, J. A. Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr. Opin. Chem. Biol. 8, 484–491 (2004)
Mahapatra, A. et al. Characterization of a Methanosarcina acetivorans mutant unable to translate UAG as pyrrolysine. Mol. Microbiol. 59, 56–66 (2006)
Hao, B. et al. Reactivity and chemical synthesis of l-pyrrolysine—the 22nd genetically encoded amino acid. Chem. Biol. 11, 1317–1324 (2004)
Soares, J. A. et al. The residue mass of l-pyrrolysine in three distinct methylamine methyltransferases. J. Biol. Chem. 280, 36962–36969 (2005)
Burke, S. A. & Krzycki, J. A. Reconstitution of monomethylamine:coenzyme M methyl transfer with a corrinoid protein and two methyltransferases purified from Methanosarcina barkeri. J. Biol. Chem. 272, 16570–16577 (1997)
Ferguson, D. J., Jr, Gorlatova, N., Grahame, D. A. & Krzycki, J. A. Reconstitution of dimethylamine:coenzyme M methyl transfer with a discrete corrinoid protein and two methyltransferases purified from Methanosarcina barkeri. J. Biol. Chem. 275, 29053–29060 (2000)
Ferguson, D. J., Jr & Krzycki, J. A. Reconstitution of trimethylamine-dependent coenzyme M methylation with the trimethylamine corrinoid protein and the isozymes of methyltransferase II from Methanosarcina barkeri. J. Bacteriol. 179, 846–852 (1997)
Burke, S. A., Lo, S. L. & Krzycki, J. A. Clustered genes encoding the methyltransferases of methanogenesis from monomethylamine. J. Bacteriol. 180, 3432–3440 (1998)
Paul, L., Ferguson, D. J. & Krzycki, J. A. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J. Bacteriol. 182, 2520–2529 (2000)
Longstaff, D. G. et al. A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine. Proc. Natl Acad. Sci. USA 104, 1021–1026 (2007)
Polycarpo, C. et al. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl Acad. Sci. USA 101, 12450–12454 (2004)
Krzycki, J. A. The direct genetic encoding of pyrrolysine. Curr. Opin. Microbiol. 8, 706–712 (2005)
Ambrogelly, A., Palioura, S. & Soll, D. Natural expansion of the genetic code. Nature Chem. Biol. 3, 29–35 (2007)
Jeong, H. et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J. Mol. Biol. 394, 644–652 (2009)
Namy, O. et al. Adding pyrrolysine to the Escherichia coli genetic code. FEBS Lett. 581, 5282–5288 (2007)
Li, W. T. et al. Specificity of pyrrolysyl-tRNA synthetase for pyrrolysine and pyrrolysine analogs. J. Mol. Biol. 385, 1156–1164 (2009)
Polycarpo, C. R. et al. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 580, 6695–6700 (2006)
Yanagisawa, T. et al. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode Nε-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem. Biol. 15, 1187–1197 (2008)
Rother, M. & Krzycki, J. A. Selenocysteine, pyrrolysine, and the unique energy metabolism of methanogenic archaea. Archaea 2010, 453642 (2010)
Frey, P. A., Hegeman, A. D. & Ruzicka, F. J. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43, 63–88 (2008)
Banerjee, R. Radical carbon skeleton rearrangements: catalysis by coenzyme B12-dependent mutases. Chem. Rev. 103, 2083–2094 (2003)
Wong, J. T. Coevolution theory of the genetic code at age thirty. Bioessays 27, 416–425 (2005)
Fekner, T., Li, X., Lee, M. M. & Chan, M. K. A pyrrolysine analogue for protein click chemistry. Angew. Chem. Int. Edn Engl. 48, 1633–1635 (2009)
Chen, P. R. et al. A facile system for encoding unnatural amino acids in mammalian cells. Angew. Chem. Int. Edn 48, 4052–4055 (2009)
Neumann, H., Peak-Chew, S. Y. & Chin, J. W. Genetically encoding Nε-acetyllysine in recombinant proteins. Nature Chem. Biol. 4, 232–234 (2008)
Xu, H. & Freitas, M. A. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics 8, 133 (2007)
Acknowledgements
This work was supported by a grant from the National Institutes of Health to K.B.G.-C. and by grants from the US Department of Energy and the National Institutes of Health to J.A.K. The authors are grateful to D. Longstaff for preparing the tagged mtmB1 version of pDLBAD.
Author information
Authors and Affiliations
Contributions
M.A.G. planned experiments, labelled and purified MtmB samples from recombinant cells, made recombinant strains, carried out immunoblotting, analysed data and prepared figures. L.Z. carried out proteolytic digestion of MtmB, determined peptide and residue masses, analysed data and helped prepare data for presentation. K.B.G.-C. oversaw data acquisition and primary data analysis. J.A.K. planned and advised on experiments, analysed and interpreted data, prepared figures and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-3 with legends and Supplementary Tables 1-8. (PDF 1319 kb)
Rights and permissions
About this article
Cite this article
Gaston, M., Zhang, L., Green-Church, K. et al. The complete biosynthesis of the genetically encoded amino acid pyrrolysine from lysine. Nature 471, 647–650 (2011). https://doi.org/10.1038/nature09918
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09918
This article is cited by
-
Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex
Communications Biology (2023)
-
Improved pyrrolysine biosynthesis through phage assisted non-continuous directed evolution of the complete pathway
Nature Communications (2021)
-
Factors in Protobiomonomer Selection for the Origin of the Standard Genetic Code
Acta Biotheoretica (2021)
-
Genome recoding strategies to improve cellular properties: mechanisms and advances
aBIOTECH (2021)
-
Improving the Microbial Production of Amino Acids: From Conventional Approaches to Recent Trends
Biotechnology and Bioprocess Engineering (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.