Abstract
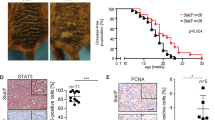
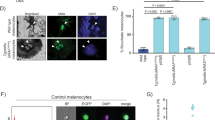
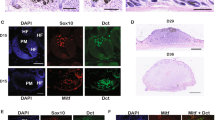
The most common mutation in human melanoma, BRAF(V600E), activates the serine/threonine kinase BRAF and causes excessive activity in the mitogen-activated protein kinase pathway1,2. BRAF(V600E) mutations are also present in benign melanocytic naevi3, highlighting the importance of additional genetic alterations in the genesis of malignant tumours. Such changes include recurrent copy number variations that result in the amplification of oncogenes4,5. For certain amplifications, the large number of genes in the interval has precluded an understanding of the cooperating oncogenic events. Here we have used a zebrafish melanoma model to test genes in a recurrently amplified region of chromosome 1 for the ability to cooperate with BRAF(V600E) and accelerate melanoma. SETDB1, an enzyme that methylates histone H3 on lysine 9 (H3K9), was found to accelerate melanoma formation significantly in zebrafish. Chromatin immunoprecipitation coupled with massively parallel DNA sequencing and gene expression analyses uncovered genes, including HOX genes, that are transcriptionally dysregulated in response to increased levels of SETDB1. Our studies establish SETDB1 as an oncogene in melanoma and underscore the role of chromatin factors in regulating tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Davies, H. et al. Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002)
Wan, P. T. et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF . Cell 116, 855–867 (2004)
Pollock, P. M. et al. High frequency of BRAF mutations in nevi. Nature Genet. 33, 19–20 (2003)
Curtin, J. A. et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 353, 2135–2147 (2005)
Garraway, L. A. et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436, 117–122 (2005)
Lin, W. M. et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 68, 664–673 (2008)
Patton, E. E. et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr. Biol. 15, 249–254 (2005)
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010)
Michaloglou, C. et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436, 720–724 (2005)
Dimri, G. P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo . Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995)
Santoriello, C. et al. Expression of H-RASV12 in a zebrafish model of Costello syndrome causes cellular senescence in adult proliferating cells. Dis. Model. Mech. 2, 56–67 (2009)
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005)
Mootha, V. K. et al. PGC-1α responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genet. 34, 267–273 (2003)
Bilodeau, S., Kagey, M. H., Frampton, G. M., Rahl, P. B. & Young, R. A. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 23, 2484–2489 (2009)
Wang, H. et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell 12, 475–487 (2003)
Fritsch, L. et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol. Cell 37, 46–56 (2010)
Dalgliesh, G. L. et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010)
van Haaften, G. et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature Genet. 41, 521–523 (2009)
Kwan, K. M. et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007)
Beroukhim, R. et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl Acad. Sci. USA 104, 20007–20012 (2007)
Nisolle, M. et al. Immunohistochemical study of the proliferation index, oestrogen receptors and progesterone receptors A and B in leiomyomata and normal myometrium during the menstrual cycle and under gonadotrophin-releasing hormone agonist therapy. Hum. Reprod. 14, 2844–2850 (1999)
Perner, S. et al. EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia 10, 298–302 (2008)
Sarraf, S. A. & Stancheva, I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell 15, 595–605 (2004)
Acknowledgements
We thank D. Harrington, R. White and Y. Zhou for discussions; C. Lawrence, I. Adatto and L.-K. Zhang for expert fish care; G. Frampton for bioinformatics assistance; and K. Kwan, C.-B. Chien, and J. Boehm for reagents. This work was supported by grants from the Damon Runyon Cancer Research Foundation (C.J.C., DRG-1855-05), the Charles A. King Trust Foundation (C.J.C.), a Young Investigator Award from the American Society of Clinical Oncology (Y.H.), the Canadian Institutes of Health Research (S.B.) and the National Institutes of Health (C.J.C., K99AR056899-02; Y.H., K08DK075432-04; R.A.Y., CA146455, HG002668; and L.I.Z., CA103846 and DK055381).
Author information
Authors and Affiliations
Contributions
C.J.C. and Y.H. contributed equally to this work and are listed alphabetically. C.J.C., Y.H. and L.I.Z. conceived the project, designed and analysed the experiments, and wrote the manuscript. C.J.C. and Y.H. performed the zebrafish experiments and contributed to the other experiments. J.J.-V. performed the tissue culture experiments. S.B. performed the ChIP-seq experiments and analysed the data. V.B., L.F., S.A.-S.-A. performed the biochemistry studies on SETDB1. L.A.J. performed the fluorescence in situ hybridization studies. T.J.H. performed the immunohistochemistry experiments. W.M.L., R.B. and C.H.M. analysed the copy number data. D.A.O. analysed the SETDB1-overexpression microarray data for WM451Lu cells. F.F. designed a database to manage and analyse tumour incidence data. C.B., C.J.B., L.T. and A.U. provided technical assistance. M.L., L.A.G. and R.A.Y. provided input into the preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
L.I.Z. is a founder and stockholder of Fate Therapeutics and a scientific adviser for Stemgent.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-18 with legends and 2 additional references. (PDF 14772 kb)
Supplementary Tables
This file contains Supplementary Tables 1-5 (Table 1 pp 1-406, Table 2 pp 407-896, Table 3 pp 897-923, Table 4 pg 924 and Table 5 pp 925-926). (PDF 24070 kb)
Rights and permissions
About this article
Cite this article
Ceol, C., Houvras, Y., Jane-Valbuena, J. et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 471, 513–517 (2011). https://doi.org/10.1038/nature09806
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09806
This article is cited by
-
Cell-type differential targeting of SETDB1 prevents aberrant CTCF binding, chromatin looping, and cis-regulatory interactions
Nature Communications (2024)
-
Histone Methyltransferase G9a Plays an Essential Role on Nicotine Preference in Zebrafish
Molecular Neurobiology (2024)
-
The functions of SET domain bifurcated histone lysine methyltransferase 1 (SETDB1) in biological process and disease
Epigenetics & Chromatin (2023)
-
Histone methyltransferase SETDB1 promotes osteogenic differentiation in osteoporosis by activating OTX2-mediated BMP-Smad and Wnt/β-catenin pathways
Human Cell (2023)
-
Current understanding of epigenetics role in melanoma treatment and resistance
Cancer Cell International (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.