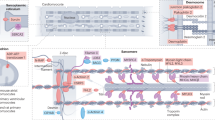
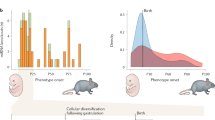
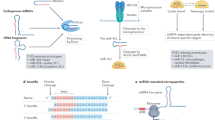
Abstract
First recognized as regulators of development in worms and fruitflies, microRNAs are emerging as pivotal modulators of mammalian cardiovascular development and disease. Individual microRNAs modulate the expression of collections of messenger RNA targets that often have related functions, thereby governing complex biological processes. The wideranging functions of microRNAs in the cardiovascular system have provided new perspectives on disease mechanisms and have revealed intriguing therapeutic targets, as well as diagnostics, for a variety of cardiovascular disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hill, J. A. & Olson, E. N. Cardiac plasticity. N. Engl. J. Med. 358, 1370–1380 (2008).
Hoffman, J. I. & Kaplan, S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 39, 1890–1900 (2002).
Bruneau, B. G. The developmental genetics of congenital heart disease. Nature 451, 943–948 (2008).
Cordes, K. R. & Srivastava, D. MicroRNA regulation of cardiovascular development. Circ. Res. 104, 724–732 (2009).
Latronico, M. V. & Condorelli, G. MicroRNAs and cardiac pathology. Nature Rev. Cardiol. 6, 419–429 (2009).
Small, E. M., Frost, R. J. & Olson, E. N. MicroRNAs add a new dimension to cardiovascular disease. Circulation 121, 1022–1032 (2010).
van Rooij, E. & Olson, E. N. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J. Clin. Invest. 117, 2369–2376 (2007).
Liu, N. & Olson, E. N. MicroRNA regulatory networks in cardiovascular development. Dev. Cell 18, 510–525 (2010).
Ikeda, S. et al. Altered microRNA expression in human heart disease. Physiol. Genomics 31, 367–373 (2007).
van Rooij, E. et al. A signature pattern of stress–responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl Acad. Sci. USA 103, 18255–18260 (2006). This important paper describes the dynamic regulation of miRNA expression during cardiac stress.
Matkovich, S. J. et al. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation 119, 1263–1271 (2009).
Thum, T. et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 116, 258–267 (2007).
Roy, S. et al. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc. Res. 82, 21–29 (2009).
van Rooij, E. et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl Acad. Sci. USA 105, 13027–13032 (2008).
Ji, R. et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of microRNA in vascular neointimal lesion formation. Circ. Res. 100, 1579–1588 (2007).
Xin, M. et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 23, 2166–2178 (2009).
Huang, Z. P., Neppl, R. L. & Wang, D. Z. MicroRNAs in cardiac remodeling and disease. J. Cardiovasc. Transl. Res. 3, 212–218 (2010).
van Rooij, E., Marshall, W. S. & Olson, E. N. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ. Res. 103, 919–928 (2008).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Filipowicz, W., Bhattacharyya, S. N. & Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Rev. Genet. 9, 102–114 (2008).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Lutter, D., Marr, C., Krumsiek, J., Lang, E. W. & Theis, F. J. Intronic microRNAs support their host genes by mediating synergistic and antagonistic regulatory effects. BMC Genomics 11, 224 (2010).
Cao, G. et al. Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways. Biochem. Biophys. Res. Commun. 396, 978–982 (2010).
Najafi-Shoushtari, S. H. et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328, 1566–1569 (2010).
Poliseno, L. et al. Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci. Signal. 3, ra29 (2010).
Barik, S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res. 36, 5232–5241 (2008).
Alvarez-Saavedra, E. & Horvitz, H. R. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20, 367–373 (2010).
Ambros, V. MicroRNAs: genetically sensitized worms reveal new secrets. Curr. Biol. 20, R598–R600 (2010).
Brenner, J. L., Jasiewicz, K. L., Fahley, A. F., Kemp, B. J. & Abbott, A. L. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans . Curr. Biol. 20, 1321–1325 (2010). This paper suggests redundant and stress-responsive roles of miRNAs, through using miRNA mutants in Dicer -deficient C. elegans.
Zhao, Y. et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129, 303–317 (2007). This paper demonstrates an important role for an miRNA in heart development by genetic deletion in mice.
Yang, B. et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2 . Nature Med. 13, 486–491 (2007).
Luo, X. et al. Down-regulation of miR-1/miR-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J. Biol. Chem. 283, 20045–20052 (2008).
Small, E. M. et al. Regulation of PI3-kinase/Akt signalling by muscle-enriched microRNA-486. Proc. Natl Acad. Sci. USA 107, 4218–4223 (2010).
Xu, N., Papagiannakopoulos, T., Pan, G., Thomson, J. A. & Kosik, K. S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137, 647–658 (2009).
Choi, W. Y., Giraldez, A. J. & Schier, A. F. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318, 271–274 (2007).
Xiao, J. et al. Novel approaches for gene-specific interference via manipulating actions of microRNAs: examination on the pacemaker channel genes HCN2 and HCN4 . J. Cell. Physiol. 212, 285–292 (2007).
Brown, B. D. & Naldini, L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nature Rev. Genet. 10, 578–585 (2009).
Ebert, M. S., Neilson, J. R. & Sharp, P. A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nature Methods 4, 721–726 (2007).
Chen, J. F. et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc. Natl Acad. Sci. USA 105, 2111–2116 (2008).
Albinsson, S. et al. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler. Thromb. Vasc. Biol. 30, 1118–1126 (2010).
Rao, P. K. et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ. Res. 105, 585–594 (2009). Deep sequencing showed that the 18 most abundant cardiac miRNAs account for more than 90% of all miRNAs in the heart.
Brown, B. D. et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nature Biotechnol. 25, 1457–1467 (2007).
Liu, N. et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl Acad. Sci. USA 104, 20844–20849 (2007).
Zhao, Y., Samal, E. & Srivastava, D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436, 214–220 (2005).
Ivey, K. N. et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell 2, 219–229 (2008).
Liu, N. et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 22, 3242–3254 (2008).
Deacon, D. C. et al. The miR-143–adducin3 pathway is essential for cardiac chamber morphogenesis. Development 137, 1887–1896 (2010).
Morton, S. U. et al. microRNA-138 modulates cardiac patterning during embryonic development. Proc. Natl Acad. Sci. USA 105, 17830–17835 (2008).
Schmidt, M. et al. EGFL7 regulates the collective migration of endothelial cells by restricting their spatial distribution. Development 134, 2913–2923 (2007).
Nicoli, S. et al. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature 464, 1196–1200 (2010).
Kuhnert, F. et al. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126 . Development 135, 3989–3993 (2008).
Wang, S. et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 15, 261–271 (2008).
Fish, J. E. et al. miR-126 regulates angiogenic signalling and vascular integrity. Dev. Cell 15, 272–284 (2008). References 52 and 53 show a crucial role for miR-126 in angiogenesis.
Small, E. M., Sutherland, L. B., Rajagopalan, R., Wang, S. & Olson, E. N. MicroRNA-218 regulates vascular patterning by modulation of Slit–Robo signaling. Circ. Res. 107, 1336–1344 (2010).
Cordes, K. R. et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460, 705–710 (2009).
Elia, L. et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 16, 1590–1598 (2009).
Boettger, T. et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J. Clin. Invest. 119, 2634–2647 (2009).
Zhao, G., Yu, D. & Weiss, M. J. MicroRNAs in erythropoiesis. Curr. Opin. Hematol. 17, 155–162 (2010).
Georgantas, R. W. III et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc. Natl Acad. Sci. USA 104, 2750–2755 (2007).
O'Carroll, D. et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 21, 1999–2004 (2007).
Lu, J. et al. MicroRNA-mediated control of cell fate in megakaryocyte–erythrocyte progenitors. Dev. Cell 14, 843–853 (2008).
Wang, Q. et al. MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood 111, 588–595 (2008).
Rasmussen, K. D. et al. The miR-144/451 locus is required for erythroid homeostasis. J. Exp. Med. 207, 1351–1358 (2010).
Johnnidis, J. B. et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451, 1125–1129 (2008).
Patrick, D. M. et al. Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3ζ. Genes Dev. 24, 1614–1619 (2010).
Yu, D. et al. miR-451 protects against erythroid oxidant stress by repressing 14-3-3ζ. Genes Dev. 24, 1620–1633 (2010). References 65 and 66 show that miR-451 is required for proper erythroid differentiation, and suggest a potential therapeutic application for targeting miR-451 for degradation.
Callis, T. E. et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 119, 2772–2786 (2009).
Thum, T. et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456, 980–984 (2008). This paper demonstrates an important role for miR-21 in cardiac remodelling using antagomir-mediated knockdown in mice.
van Rooij, E. et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579 (2007). The first paper to show a role for an miRNA, miR-208a, in the control of cardiac remodelling, using a genetic knockout.
Care, A. et al. MicroRNA-133 controls cardiac hypertrophy. Nature Med. 13, 613–618 (2007).
van Rooij, E. et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell 17, 662–673 (2009).
Molkentin, J. D. et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93, 215–228 (1998).
Lin, Z. et al. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc. Natl Acad. Sci. USA 106, 12103–12108 (2009).
Rane, S. et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1α and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ. Res. 104, 879–886 (2009).
Patrick, D. M. et al. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J. Clin. Invest. 120, 3912–3916 (2010).
Fleissner, F. et al. Asymmetric dimethylarginine impairs angiogenic progenitor cell function in patients with coronary artery disease through a microRNA-21-dependent mechanism. Circ. Res. 107, 138–143 (2010).
Bonauer, A. et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 324, 1710–1713 (2009).
van Solingen, C. et al. Antagomir-mediated silencing of endothelial cell specific microRNA-126 impairs ischemia-induced angiogenesis. J. Cell. Mol. Med. 13, 1577–1585 (2009).
Zernecke, A. et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci. Signal. 2, ra81 (2009).
Harris, T. A., Yamakuchi, M., Ferlito, M., Mendell, J. T. & Lowenstein, C. J. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl Acad. Sci. USA 105, 1516–1521 (2008).
Cheng, Y. et al. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ. Res. 105, 158–166 (2009).
Saunders, M. A., Liang, H. & Li, W. H. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl Acad. Sci. USA 104, 3300–3305 (2007).
Chen, K. & Rajewsky, N. Natural selection on human microRNA binding sites inferred from SNP data. Nature Genet. 38, 1452–1456 (2006).
Clop, A. et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nature Genet. 38, 813–818 (2006).
Sethupathy, P. et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am. J. Hum. Genet. 81, 405–413 (2007).
Schipper, M. E., van Kuik, J., de Jonge, N., Dullens, H. F. & de Weger, R. A. Changes in regulatory microRNA expression in myocardium of heart failure patients on left ventricular assist device support. J. Heart Lung Transplant. 27, 1282–1285 (2008).
Voellenkle, C. et al. MicroRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol. Genomics 42, 420–426 (2010).
Fichtlscherer, S. et al. Circulating microRNAs in patients with coronary artery disease. Circ. Res. 107, 677–684 (2010).
Ji, X. et al. Plasma miR-208 as a biomarker of myocardial injury. Clin. Chem. 55, 1944–1949 (2009).
Elmen, J. et al. LNA-mediated microRNA silencing in non-human primates. Nature 452, 896–899 (2008).
Lanford, R. E. et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327, 198–201 (2010). The first report of therapeutically targeting an miRNA for the treatment of a disease in non-human primates.
Acknowledgements
We apologize to all colleagues whose work could not be cited owing to space restrictions. We thank J. Cabrera for artwork and J. Brown for editorial assistance. E.N.O. was supported by grants from the National Institutes of Health, the Donald W. Reynolds Center for Clinical Cardiovascular Research, the Robert A. Welch Foundation, the Fondation Leducq's Transatlantic Network for Excellence in Cardiovascular Research Program, the American Heart Association and the Jon Holden DeHaan Foundation. E.M.S. was supported by a scientist development grant from the American Heart Association.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
E.N.O. holds equity in miRagen Therapeutics, which is developing miRNA-based therapies for muscle disease.
Additional information
Reprints and permissions information is available at http://www.nature.com/reprints.
Rights and permissions
About this article
Cite this article
Small, E., Olson, E. Pervasive roles of microRNAs in cardiovascular biology. Nature 469, 336–342 (2011). https://doi.org/10.1038/nature09783
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09783
This article is cited by
-
MiR-181a protects the heart against myocardial infarction by regulating mitochondrial fission via targeting programmed cell death protein 4
Scientific Reports (2024)
-
Integrated small RNA, mRNA and protein omics reveal a miRNA network orchestrating metabolic maturation of the developing human heart
BMC Genomics (2023)
-
The KLF7/PFKL/ACADL axis modulates cardiac metabolic remodelling during cardiac hypertrophy in male mice
Nature Communications (2023)
-
Exercise and chronic kidney disease: potential mechanisms underlying the physiological benefits
Nature Reviews Nephrology (2023)
-
miR-322 promotes the differentiation of embryonic stem cells into cardiomyocytes
Functional & Integrative Genomics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.