Abstract
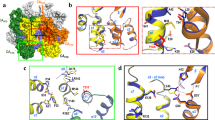
The mature capsids of human immunodeficiency virus type 1 (HIV-1) and other retroviruses are fullerene shells, composed of the viral CA protein, that enclose the viral genome and facilitate its delivery into new host cells1. Retroviral CA proteins contain independently folded amino (N)- and carboxy (C)-terminal domains (NTD and CTD) that are connected by a flexible linker2,3,4. The NTD forms either hexameric or pentameric rings, whereas the CTD forms symmetric homodimers that connect the rings into a hexagonal lattice3,5,6,7,8,9,10,11,12,13. We previously used a disulphide crosslinking strategy to enable isolation and crystallization of soluble HIV-1 CA hexamers11,14. Here we use the same approach to solve the X-ray structure of the HIV-1 CA pentamer at 2.5 Å resolution. Two mutant CA proteins with engineered disulphides at different positions (P17C/T19C and N21C/A22C) converged onto the same quaternary structure, indicating that the disulphide-crosslinked proteins recapitulate the structure of the native pentamer. Assembly of the quasi-equivalent hexamers and pentamers requires remarkably subtle rearrangements in subunit interactions, and appears to be controlled by an electrostatic switch that favours hexamers over pentamers. This study completes the gallery of substructures describing the components of the HIV-1 capsid and enables atomic-level modelling of the complete capsid. Rigid-body rotations around two assembly interfaces appear sufficient to generate the full range of continuously varying lattice curvature in the fullerene cone.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
The coordinates and structure factors are deposited in Protein Data Bank under accession numbers 3P05 (N21C/A22C-stabilized pentamer) and 3P0A (P17C/T19C-stabilized pentamer). To reflect the limited resolution of the second structure properly, only Cα coordinates are deposited.
References
Ganser-Pornillos, B. K., Yeager, M. & Sundquist, W. I. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18, 203–217 (2008)
Gitti, R. K. et al. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273, 231–235 (1996)
Gamble, T. R. et al. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278, 849–853 (1997)
Berthet-Colominas, C. et al. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 18, 1124–1136 (1999)
Worthylake, D. K., Wang, H., Yoo, S., Sundquist, W. I. & Hill, C. P. Structures of the HIV-1 capsid protein dimerization domain at 2.6 Å resolution. Acta Crystallogr. D 55, 85–92 (1999)
Li, S., Hill, C. P., Sundquist, W. I. & Finch, J. T. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407, 409–413 (2000)
Mortuza, G. B. et al. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 431, 481–485 (2004)
Ganser-Pornillos, B. K., Cheng, A. & Yeager, M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell 131, 70–79 (2007)
Cardone, G., Purdy, J. G., Cheng, N., Craven, R. C. & Steven, A. C. Visualization of a missing link in retrovirus capsid assembly. Nature 457, 694–698 (2009)
Bailey, G. D., Hyun, J. K., Mitra, A. K. & Kingston, R. L. Proton-linked dimerization of a retroviral capsid protein initiates capsid assembly. Structure 17, 737–748 (2009)
Pornillos, O. et al. X-ray structures of the hexameric building block of the HIV capsid. Cell 137, 1282–1292 (2009)
Byeon, I. J. et al. Structural convergence between cryoEM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell 139, 780–790 (2009)
Hyun, J. K., Radjainia, M., Kingston, R. L. & Mitra, A. K. Proton-driven assembly of the Rous sarcoma virus capsid protein results in the formation of icosahedral particles. J. Biol. Chem. 285, 15056–15064 (2010)
Pornillos, O., Ganser-Pornillos, B. K., Banumathi, S., Hua, Y. & Yeager, M. Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus. J. Mol. Biol. 401, 985–995 (2010)
Ganser, B. K., Li, S., Klishko, V. Y., Finch, J. T. & Sundquist, W. I. Assembly and analysis of conical models for the HIV-1 core. Science 283, 80–83 (1999)
Jin, Z., Jin, L., Peterson, D. L. & Lawson, C. L. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J. Mol. Biol. 286, 83–93 (1999)
Heymann, J. B., Butan, C., Winkler, D. C., Craven, R. C. & Steven, A. C. Irregular and semi-regular polyhedral models for Rous sarcoma virus cores. Comput. Math. Methods Med. 9, 197–210 (2008)
Caspar, D. L. & Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 27, 1–24 (1962)
Ganser-Pornillos, B. K., von Schwedler, U. K., Stray, K. M., Aiken, C. & Sundquist, W. I. Assembly properties of the human immunodeficiency virus type 1 CA protein. J. Virol. 78, 2545–2552 (2004)
Ehrlich, L. S., Agresta, B. E. & Carter, C. A. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro . J. Virol. 66, 4874–4883 (1992)
Campbell, S. & Vogt, V. M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69, 6487–6497 (1995)
Gross, I., Hohenberg, H. & Kräusslich, H. G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249, 592–600 (1997)
Ternois, F., Sticht, J., Duquerroy, S., Kräusslich, H. G. & Rey, F. A. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nature Struct. Mol. Biol. 12, 678–682 (2005)
Ivanov, D. et al. Domain-swapped dimerization of the HIV-1 capsid C-terminal domain. Proc. Natl Acad. Sci. USA 104, 4353–4358 (2007)
Alcaraz, L. A., del Alamo, M., Barrera, F. N., Mateu, M. G. & Neira, J. L. Flexibility in HIV-1 assembly subunits: solution structure of the monomeric C-terminal domain of the capsid protein. Biophys. J. 93, 1264–1276 (2007)
Bartonova, V. et al. Residues in the HIV-1 capsid assembly inhibitor binding site are essential for maintaining the assembly-competent quaternary structure of the capsid protein. J. Biol. Chem. 283, 32024–32033 (2008)
Wong, H. C., Shin, R. & Krishna, N. R. Solution structure of a double mutant of the carboxy-terminal dimerization domain of the HIV-1 capsid protein. Biochemistry 47, 2289–2297 (2008)
von Schwedler, U. K., Stray, K. M., Garrus, J. E. & Sundquist, W. I. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77, 5439–5450 (2003)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30, 1022–1025 (1997)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Reddy, V. et al. Effective electron-density map improvement and structure validation on a Linux multi-CPU web cluster: the TB Structural Genomics Consortium Bias Removal Web Service. Acta Crystallogr. D 59, 2200–2210 (2003)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Kleywegt, G. J. & Jones, T. A. xdlMAPMAN and xdlDATAMAN – programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr. D 52, 826–828 (1996)
Vaguine, A. A., Richelle, J. & Wodak, S. J. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D 55, 191–205 (1999)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Wriggers, W. & Schulten, K. Protein domain movements: detection of rigid domains and visualization of hinges in comparisons of atomic coordinates. Proteins 29, 1–14 (1997)
Acknowledgements
This study was funded by grants from the US National Institutes of Health to M.Y. (R01-GM066087 and P50-GM082545). X-ray diffraction data were collected at beamlines 22-BM and 22-ID at the Advanced Photon Source, Argonne National Laboratory. Initial crystal screening was performed with the assistance of S. Banumathi through the Collaborative Crystallography Program, Lawrence Berkeley National Laboratory at the Advanced Light Source. We thank J. E. Johnson and D. Borek for crystallographic advice; Y. Hua for assistance with molecular biology experiments; and I. A. Wilson, C. P. Hill and W. I. Sundquist for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
All authors designed/performed the experiments, analysed the data and wrote the manuscript. O.P. performed the computational aspects of crystallographic structure determination.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Table 1 and Supplementary Figures 1-5 with legends. (PDF 5445 kb)
Rights and permissions
About this article
Cite this article
Pornillos, O., Ganser-Pornillos, B. & Yeager, M. Atomic-level modelling of the HIV capsid. Nature 469, 424–427 (2011). https://doi.org/10.1038/nature09640
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09640
This article is cited by
-
Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization
Nature Communications (2023)
-
Rotten to the core: antivirals targeting the HIV-1 capsid core
Retrovirology (2021)
-
The HIV-1 capsid and reverse transcription
Retrovirology (2021)
-
HIV-1 capsid exploitation of the host microtubule cytoskeleton during early infection
Retrovirology (2021)
-
Macromolecular crystallization: basics and advanced methodologies
Journal of the Iranian Chemical Society (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.