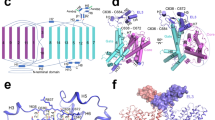
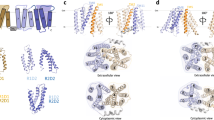
Abstract
Membrane co-transport proteins that use a five-helix inverted repeat motif have recently emerged as one of the largest structural classes of secondary active transporters1,2. However, despite many structural advances there is no clear evidence of how ion and substrate transport are coupled. Here we report a comprehensive study of the sodium/galactose transporter from Vibrio parahaemolyticus (vSGLT), consisting of molecular dynamics simulations, biochemical characterization and a new crystal structure of the inward-open conformation at a resolution of 2.7 Å. Our data show that sodium exit causes a reorientation of transmembrane helix 1 that opens an inner gate required for substrate exit, and also triggers minor rigid-body movements in two sets of transmembrane helical bundles. This cascade of events, initiated by sodium release, ensures proper timing of ion and substrate release. Once set in motion, these molecular changes weaken substrate binding to the transporter and allow galactose readily to enter the intracellular space. Additionally, we identify an allosteric pathway between the sodium-binding sites, the unwound portion of transmembrane helix 1 and the substrate-binding site that is essential in the coupling of co-transport.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Abramson, J. & Wright, E. M. Structure and function of Na+-symporters with inverted repeats. Curr. Opin. Struct. Biol. 19, 425–432 (2009)
Krishnamurthy, H., Piscitelli, C. L. & Gouaux, E. Unlocking the molecular secrets of sodium-coupled transporters. Nature 459, 347–355 (2009)
Jardetzky, O. Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966)
Wright, E. M., Hirayama, B. A. & Loo, D. F. Active sugar transport in health and disease. J. Intern. Med. 261, 32–43 (2007)
Isaji, M. Sodium-glucose cotransporter inhibitors for diabetes. Curr. Opin. Investig. Drugs 8, 285–292 (2007)
Faham, S. et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321, 810–814 (2008)
Weyand, S. et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science 322, 709–713 (2008)
Ressl, S., Terwisscha van Scheltinga, A. C., Vonrhein, C., Ott, V. & Ziegler, C. Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature 458, 47–52 (2009)
Fang, Y. et al. Structure of a prokaryotic virtual proton pump at 3.2 Å resolution. Nature 460, 1040–1043 (2009)
Gao, X. et al. Structure and mechanism of an amino acid antiporter. Science 324, 1565–1568 (2009)
Shaffer, P. L., Goehring, A., Shankaranarayanan, A. & Gouaux, E. Structure and mechanism of a Na+-independent amino acid transporter. Science 325, 1010–1014 (2009)
Yamashita, A., Singh, S. K., Kawate, T., Jin, Y. & Gouaux, E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215–223 (2005)
Forrest, L. R. & Rudnick, G. The rocking bundle: a mechanism for ion-coupled solute flux by symmetrical transporters. Physiology (Bethesda) 24, 377–386 (2009)
Zhou, Z. et al. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science 317, 1390–1393 (2007)
Gao, X. et al. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature 463, 828–832 (2010)
Singh, S. K., Piscitelli, C. L., Yamashita, A. & Gouaux, E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science 322, 1655–1661 (2008)
Shimamura, T. et al. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science 328, 470–473 (2010)
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008)
Li, J. & Tajkhorshid, E. Ion-releasing state of a secondary membrane transporter. Biophys. J. 97, L29–L31 (2009)
Quick, M., Loo, D. D. & Wright, E. M. Neutralization of a conserved amino acid residue in the human Na+/glucose transporter (hSGLT1) generates a glucose-gated H+ channel. J. Biol. Chem. 276, 1728–1734 (2001)
Kumar, S., Bouzida, D., Swendsen, R. H., Kollman, P. A. & Rosenberg, J. M. The weighted histogram analysis method for free-energy calculations on biomolecules. 1. The method. J. Comput. Chem. 13, 1011–1021 (1992)
Yernool, D., Boudker, O., Jin, Y. & Gouaux, E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii . Nature 431, 811–818 (2004)
Hunte, C. et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435, 1197–1202 (2005)
Lomize, A. L., Pogozheva, I. D., Lomize, M. A. & Mosberg, H. I. Positioning of proteins in membranes: a computational approach. Protein Sci. 15, 1318–1333 (2006)
Kale, L. et al. NAMD2: Greater scalability for parallel molecular dynamics. J. Comput. Phys. 151, 283–312 (1999)
Sali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993)
MacKerell, A. D., Jr, Feig, M. & Brooks, C. L., III Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 126, 698–699 (2004)
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996)
Ryckaert, J. P., Ciccotti, G. & Berendsen, H. J. C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977)
Turk, E. et al. Molecular characterization of Vibrio parahaemolyticus vSGLT: a model for sodium-coupled sugar cotransporters. J. Biol. Chem. 275, 25711–25716 (2000)
Turk, E., Gasymov, O. K., Lanza, S., Horwitz, J. & Wright, E. M. A reinvestigation of the secondary structure of functionally active vSGLT, the Vibrio sodium/galactose cotransporter. Biochemistry 45, 1470–1479 (2006)
Veenstra, M., Lanza, S., Hirayama, B. A., Turk, E. & Wright, E. M. Local conformational changes in the Vibrio Na+/galactose cotransporter. Biochemistry 43, 3620–3627 (2004)
Otwinowski, Z. & Minor, W. in Macromolecular Crystallography (eds Abelson, J. N., Simon, M. I., Carter, C. W. Jr & Sweet, R. M. ) 307–326 (Methods in Enzymology 276, Academic, 1997)
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006)
Qian, B. et al. High-resolution structure prediction and the crystallographic phase problem. Nature 450, 259–264 (2007)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11, 53–55 (2004)
Joosten, R. P., Womack, T., Vriend, G. & Bricogne, G. Re-refinement from deposited X-ray data can deliver improved models for most PDB entries. Acta Crystallogr. D 65, 176–185 (2009)
DeLano, W. L. PyMOL Molecular Viewer 〈http://www.pymol.org〉 (2002)
Acknowledgements
We thank T. Vondriska and K. Philipson as well as members of the Abramson, Wright and Grabe labs for useful discussions and for reading the manuscript. We would also like to thank S. Faham for contributions at the early stages of this work, S. Iwata for advance release of the Mhp1 coordinates (Protein Data Bank ID, 2X79), and R. Roskies for assistance with the computations. Simulations were carried out through a TeraGrid grant at the Pittsburgh Supercomputing Center and the Texas Advanced Computing Center. This work was supported by NIH grants GM078844 (J.A.), RGY0069 (J.A.) and DK19567 (E.M.W.), and a grant from the Human Frontier Science Program (J.A.). M.G. is an Alfred P. Sloan Research Fellow.
Author information
Authors and Affiliations
Contributions
Experiments were carried out and diffraction data collected by A.W., V.C. and J.A. Simulations were carried out by S.C. Data were analysed and the manuscript was prepared by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-6 with legends, Supplementary Table 1 and a legend for Supplementary Movie 1. (PDF 2560 kb)
Supplementary Movie 1
This movie shows a 200 ns unrestrained MD simulation of galactose exiting from the sodium glucose transporter from Vibrio parahaemolyticus (see Supplementary Information file for full legend. (MOV 7132 kb)
Rights and permissions
About this article
Cite this article
Watanabe, A., Choe, S., Chaptal, V. et al. The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature 468, 988–991 (2010). https://doi.org/10.1038/nature09580
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09580
This article is cited by
-
Antidiabetic potential of Lysiphyllum strychnifolium (Craib) A. Schmitz compounds in human intestinal epithelial Caco-2 cells and molecular docking-based approaches
BMC Complementary Medicine and Therapies (2022)
-
Novel biallelic variants expand the SLC5A6-related phenotypic spectrum
European Journal of Human Genetics (2022)
-
Molecular characterization and nutritional regulation of sodium-dependent glucose cotransporter 1 (Sglt1) in blunt snout bream (Megalobrama amblycephala)
Scientific Reports (2021)
-
Sodium-coupled glucose transport, the SLC5 family, and therapeutically relevant inhibitors: from molecular discovery to clinical application
Pflügers Archiv - European Journal of Physiology (2020)
-
Plant glucose transporter structure and function
Pflügers Archiv - European Journal of Physiology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.