Abstract
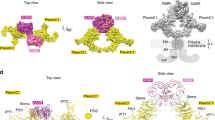
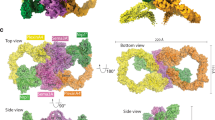
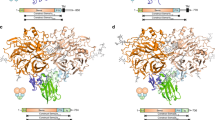
Cell-cell signalling of semaphorin ligands through interaction with plexin receptors is important for the homeostasis and morphogenesis of many tissues and is widely studied for its role in neural connectivity, cancer, cell migration and immune responses1. SEMA4D and Sema6A exemplify two diverse vertebrate, membrane-spanning semaphorin classes (4 and 6) that are capable of direct signalling through members of the two largest plexin classes, B and A, respectively2,3. In the absence of any structural information on the plexin ectodomain or its interaction with semaphorins the extracellular specificity and mechanism controlling plexin signalling has remained unresolved. Here we present crystal structures of cognate complexes of the semaphorin-binding regions of plexins B1 and A2 with semaphorin ectodomains (human PLXNB11–2–SEMA4Decto and murine PlxnA21–4–Sema6Aecto), plus unliganded structures of PlxnA21–4 and Sema6Aecto. These structures, together with biophysical and cellular assays of wild-type and mutant proteins, reveal that semaphorin dimers independently bind two plexin molecules and that signalling is critically dependent on the avidity of the resulting bivalent 2:2 complex (monomeric semaphorin binds plexin but fails to trigger signalling). In combination, our data favour a cell-cell signalling mechanism involving semaphorin-stabilized plexin dimerization, possibly followed by clustering, which is consistent with previous functional data. Furthermore, the shared generic architecture of the complexes, formed through conserved contacts of the amino-terminal seven-bladed β-propeller (sema) domains of both semaphorin and plexin, suggests that a common mode of interaction triggers all semaphorin–plexin based signalling, while distinct insertions within or between blades of the sema domains determine binding specificity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Kruger, R. P., Aurandt, J. & Guan, K. L. Semaphorins command cells to move. Nature Rev. Mol. Cell Biol. 6, 789–800 (2005)
Tamagnone, L. et al. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell 99, 71–80 (1999)
Suto, F. et al. Interactions between plexin-A2, plexin-A4, and semaphorin 6A control lamina-restricted projection of hippocampal mossy fibers. Neuron 53, 535–547 (2007)
Kolodkin, A. L., Matthes, D. J. & Goodman, C. S. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell 75, 1389–1399 (1993)
Love, C. A. et al. The ligand-binding face of the semaphorins revealed by the high-resolution crystal structure of SEMA4D. Nature Struct. Biol. 10, 843–848 (2003)
Antipenko, A. et al. Structure of the semaphorin-3A receptor binding module. Neuron 39, 589–598 (2003)
Winberg, M. L. et al. Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell 95, 903–916 (1998)
Capparuccia, L. & Tamagnone, L. Semaphorin signaling in cancer cells and in cells of the tumor microenvironment–two sides of a coin. J. Cell Sci. 122, 1723–1736 (2009)
Korostylev, A. et al. A functional role for semaphorin 4D/plexin B1 interactions in epithelial branching morphogenesis during organogenesis. Development 135, 3333–3343 (2008)
Kerjan, G. et al. The transmembrane semaphorin Sema6A controls cerebellar granule cell migration. Nature Neurosci. 8, 1516–1524 (2005)
Renaud, J. et al. Plexin-A2 and its ligand, Sema6A, control nucleus-centrosome coupling in migrating granule cells. Nature Neurosci. 11, 440–449 (2008)
He, H., Yang, T., Terman, J. R. & Zhang, X. Crystal structure of the plexin A3 intracellular region reveals an autoinhibited conformation through active site sequestration. Proc. Natl Acad. Sci. USA 106, 15610–15615 (2009)
Tong, Y. et al. Structure and function of the intracellular region of the plexin-B1 transmembrane receptor. J. Biol. Chem. 284, 35962–35972 (2009)
Takahashi, T. & Strittmatter, S. M. Plexina1 autoinhibition by the plexin sema domain. Neuron 29, 429–439 (2001)
Takahashi, T. et al. Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell 99, 59–69 (1999)
Oinuma, I., Katoh, H. & Negishi, M. Molecular dissection of the semaphorin 4D receptor plexin-B1-stimulated R-Ras GTPase-activating protein activity and neurite remodeling in hippocampal neurons. J. Neurosci. 24, 11473–11480 (2004)
Tong, Y. et al. Binding of Rac1, Rnd1, and RhoD to a novel Rho GTPase interaction motif destabilizes dimerization of the plexin-B1 effector domain. J. Biol. Chem. 282, 37215–37224 (2007)
Gherardi, E., Love, C. A., Esnouf, R. M. & Jones, E. Y. The sema domain. Curr. Opin. Struct. Biol. 14, 669–678 (2004)
Stamos, J., Lazarus, R. A., Yao, X., Kirchhofer, D. & Wiesmann, C. Crystal structure of the HGF β-chain in complex with the Sema domain of the Met receptor. EMBO J. 23, 2325–2335 (2004)
Niemann, H. H. et al. Structure of the human receptor tyrosine kinase met in complex with the Listeria invasion protein InlB. Cell 130, 235–246 (2007)
Koppel, A. M., Feiner, L., Kobayashi, H. & Raper, J. A. A 70 amino acid region within the semaphorin domain activates specific cellular response of semaphorin family members. Neuron 19, 531–537 (1997)
Müller, K. M., Arndt, K. M. & Pluckthun, A. Model and simulation of multivalent binding to fixed ligands. Anal. Biochem. 261, 149–158 (1998)
Klostermann, A., Lohrum, M., Adams, R. H. & Puschel, A. W. The chemorepulsive activity of the axonal guidance signal semaphorin D requires dimerization. J. Biol. Chem. 273, 7326–7331 (1998)
Koppel, A. M. & Raper, J. A. Collapsin-1 covalently dimerizes, and dimerization is necessary for collapsing activity. J. Biol. Chem. 273, 15708–15713 (1998)
Turner, L. J. & Hall, A. Plexin-induced collapse assay in COS cells. Methods Enzymol. 406, 665–676 (2006)
Merte, J. et al. A forward genetic screen in mice identifies Sema3A(K108N), which binds to neuropilin-1 but cannot signal. J. Neurosci. 30, 5767–5775 (2010)
Liu, H. et al. Structural basis of Semaphorin-Plexin recognition and viral mimicry from Sema7A and A39R complexes with PlexinC1. Cell 142, 749–761 (2010)
Toyofuku, T. et al. FARP2 triggers signals for Sema3A-mediated axonal repulsion. Nature Neurosci. 8, 1712–1719 (2005)
Gherardi, E. et al. Structural basis of hepatocyte growth factor/scatter factor and MET signalling. Proc. Natl Acad. Sci. USA 103, 4046–4051 (2006)
Aricescu, A. R., Lu, W. & Jones, E. Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D 62, 1243–1250 (2006)
Chang, V. T. et al. Glycoprotein structural genomics: solving the glycosylation problem. Structure 15, 267–273 (2007)
Reeves, P. J., Callewaert, N., Contreras, R. & Khorana, H. G. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl Acad. Sci. USA 99, 13419–13424 (2002)
Walter, T. S. et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. D 61, 651–657 (2005)
Mayo, C. J. et al. Benefits of automated crystallization plate tracking, imaging, and analysis. Structure 13, 175–182 (2005)
Otwinowski, Z. & Minor, W. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Leslie, A. G. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography 26, (1992)
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006)
CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Perrakis, A., Morris, R. & Lamzin, V. S. Automated protein model building combined with iterative structure refinement. Nature Struct. Biol. 6, 458–463 (1999)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 62, 1002–1011 (2006)
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D 60, 2210–2221 (2004)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
Stuart, D. I., Levine, M., Muirhead, H. & Stammers, D. K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 Å. J. Mol. Biol. 134, 109–142 (1979)
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007)
Gouet, P., Courcelle, E., Stuart, D. I. & Metoz, F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 (1999)
Landau, M. et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302 (2005)
O’Callaghan C. A. et al. BirA enzyme: production and application in the study of membrane receptor-ligand interactions by site-specific biotinylation. Anal. Biochem. 266, 9–15 (1999)
Trombetta, E. S. & Parodi, A. J. Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 19, 649–676 (2003)
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 (2000)
Acknowledgements
We thank the staff of European Synchrotron Radiation Facility beamline ID 23-1 and Diamond beamline I03 for assistance with data collection, the Molecular Cytogenetics and Microscopy Core facility of the Wellcome Trust Centre for Human Genetics, T. S. Walter for help with crystallization, G. Sutton for help with MALS experiments, A.F. Sonnen for help with AUC experiments, W. Lu for help with tissue culture, J. M. Grimes for assistance with figures and A. R. Aricescu and D. I. Stuart for critical reading of the manuscript. This work was funded by Cancer Research UK and the UK Medical Research Council. B.J.C.J. is funded by the Human Frontier Science Program, K.J.M. by a Science Foundation Ireland grant, C.H.B. and C.S. by the Wellcome Trust and E.Y.J. by Cancer Research UK.
Author information
Authors and Affiliations
Contributions
All authors contributed to the design of the project, data analysis and preparation of the manuscript. R.A.R. cloned, purified and performed SPR and MALS experiments on SEMA4D and PLXNB1 and crystallized and solved its complex structure. C.S. helped with the PLNXB1–SEMA4D complex structure solution. B.J.C.J. cloned, purified and performed SPR, AUC and MALS experiments on Sema6A and PlxnA2 and crystallized and solved the individual and complex structures. C.H.B. did the collapse assays, purified and performed AUC experiments on PLXNB1cyto and helped with other AUC experiments. F.P.-B. performed the granule cell migration assays.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains a Supplementary Table and Supplementary Figures 1-10 with legends. (PDF 9972 kb)
Rights and permissions
About this article
Cite this article
Janssen, B., Robinson, R., Pérez-Brangulí, F. et al. Structural basis of semaphorin–plexin signalling. Nature 467, 1118–1122 (2010). https://doi.org/10.1038/nature09468
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09468
This article is cited by
-
FUT8-mediated aberrant N-glycosylation of SEMA7A promotes head and neck squamous cell carcinoma progression
International Journal of Oral Science (2024)
-
Neurite outgrowth deficits caused by rare PLXNB1 mutation in pediatric bipolar disorder
Molecular Psychiatry (2023)
-
SEMA4D/PlexinB1 promotes AML progression via activation of PI3K/Akt signaling
Journal of Translational Medicine (2022)
-
Cryo-EM structure of the PlexinC1/A39R complex reveals inter-domain interactions critical for ligand-induced activation
Nature Communications (2020)
-
A comprehensive review of tumor proliferative and suppressive role of semaphorins and therapeutic approaches
Biophysical Reviews (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.