Abstract
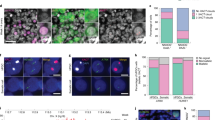
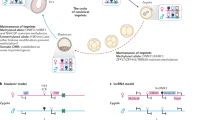
Two forms of X-chromosome inactivation (XCI) ensure the selective silencing of female sex chromosomes during mouse embryogenesis. Imprinted XCI begins with the detection of Xist RNA expression on the paternal X chromosome (Xp) at about the four-cell stage of embryonic development. In the embryonic tissues of the inner cell mass, a random form of XCI occurs in blastocysts that inactivates either Xp or the maternal X chromosome (Xm)1,2. Both forms of XCI require the non-coding Xist RNA that coats the inactive X chromosome from which it is expressed. Xist has crucial functions in the silencing of X-linked genes, including Rnf12 (refs 3, 4) encoding the ubiquitin ligase RLIM (RING finger LIM-domain-interacting protein). Here we show, by targeting a conditional knockout of Rnf12 to oocytes where RLIM accumulates to high levels, that the maternal transmission of the mutant X chromosome (Δm) leads to lethality in female embryos as a result of defective imprinted XCI. We provide evidence that in Δm female embryos the initial formation of Xist clouds and Xp silencing are inhibited. In contrast, embryonic stem cells lacking RLIM are able to form Xist clouds and silence at least some X-linked genes during random XCI. These results assign crucial functions to the maternal deposit of Rnf12/RLIM for the initiation of imprinted XCI.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Heard, E. & Disteche, C. M. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 20, 1848–1867 (2006)
Payer, B. & Lee, J. T. X chromosome dosage compensation: how mammals keep the balance. Annu. Rev. Genet. 42, 733–772 (2008)
Patrat, C. et al. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc. Natl Acad. Sci. USA 106, 5198–5203 (2009)
Kalantry, S., Purushothaman, S., Bowen, R. B., Starmer, J. & Magnuson, T. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature 460, 647–651 (2009)
Bach, I. et al. RLIM inhibits functional activity of LIM homeodomain transcription factors via recruitment of the histone deacetylase complex. Nature Genet. 22, 394–399 (1999)
Ostendorff, H. P. et al. Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature 416, 99–103 (2002)
Gungor, C. et al. Proteasomal selection of multiprotein complexes recruited by LIM homeodomain transcription factors. Proc. Natl Acad. Sci. USA 104, 15000–15005 (2007)
Johnsen, S. A. et al. Regulation of estrogen-dependent transcription by the LIM cofactors CLIM and RLIM in breast cancer. Cancer Res. 69, 128–136 (2009)
Ostendorff, H. P. et al. Functional characterization of the gene encoding RLIM, the corepressor of LIM homeodomain factors. Genomics 69, 120–130 (2000)
Ostendorff, H. P. et al. Dynamic expression of LIM cofactors in the developing mouse neural tube. Dev. Dyn. 235, 786–791 (2006)
Wagner, K. U. et al. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 10, 545–553 (2001)
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet. 21, 70–71 (1999)
Oshima, R. G., Howe, W. E., Klier, F. G., Adamson, E. D. & Shevinsky, L. H. Intermediate filament protein synthesis in preimplantation murine embryos. Dev. Biol. 99, 447–455 (1983)
Feinberg, R. F. et al. Plasminogen activator inhibitor types 1 and 2 in human trophoblasts. PAI-1 is an immunocytochemical marker of invading trophoblasts. Lab. Invest. 61, 20–26 (1989)
Beck, F., Erler, T., Russell, A. & James, R. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev. Dyn. 204, 219–227 (1995)
Marahrens, Y., Panning, B., Dausman, J., Strauss, W. & Jaenisch, R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11, 156–166 (1997)
Plath, K. et al. Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131–135 (2003)
Barlow, P. W. & Sherman, M. I. The biochemistry of differentiation of mouse trophoblast: studies on polyploidy. J. Embryol. Exp. Morphol. 27, 447–465 (1972)
Stavropoulos, N., Lu, N. & Lee, J. T. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc. Natl Acad. Sci. USA 98, 10232–10237 (2001)
Lee, J. T. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell 103, 17–27 (2000)
Hall, L. L. et al. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc. Natl Acad. Sci. USA 99, 8677–8682 (2002)
Okamoto, I., Otte, A. P., Allis, C. D., Reinberg, D. & Heard, E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303, 644–649 (2004)
Sun, B. K., Deaton, A. M. & Lee, J. T. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell 21, 617–628 (2006)
Jonkers, I. et al. RNF12 Is an X-encoded dose-dependent activator of X chromosome inactivation. Cell 139, 999–1011 (2009)
Nusslein-Volhard, C., Frohnhofer, H. G. & Lehmann, R. Determination of anteroposterior polarity in Drosophila . Science 238, 1675–1681 (1987)
Letterio, J. J. et al. Maternal rescue of transforming growth factor-β1 null mice. Science 264, 1936–1938 (1994)
Guidi, C. J. et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 21, 3598–3603 (2001)
Tursun, B. et al. The ubiquitin ligase Rnf6 regulates local LIM kinase 1 levels in axonal growth cones. Genes Dev. 19, 2307–2319 (2005)
Chung, Y. et al. Embryonic and extraembryonic stem cell lines derived from single mouse blastomeres. Nature 439, 216–219 (2006)
Wagner, K. U. et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 25, 4323–4330 (1997)
Novik, E. I., Maguire, T. J., Orlova, K., Schloss, R. S. & Yarmush, M. L. Embryoid body-mediated differentiation of mouse embryonic stem cells along a hepatocyte lineage: insights from gene expression profiles. Tissue Eng. 12, 1515–1525 (2006)
Chung, Y. & Becker, S. Embryonic stem cells using nuclear transfer. Methods Enzymol. 418, 135–147 (2006)
Chung, Y. et al. Human embryonic stem cell lines generated without embryo destruction. Cell Stem Cell 2, 113–117 (2008)
Nagy, A., Gerstenstein, M., Vintersten, K. & Behringer, R. Manipulating the Mouse Embryo (Cold Spring Harbor Laboratory Press, 2003)
Marks, H. et al. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 19, 1361–1373 (2009)
Panning, B. X inactivation in mouse ES cells: histone modifications and FISH. Methods Enzymol. 376, 419–428 (2004)
Clemson, C. M., Hall, L. L., Byron, M., McNeil, J. & Lawrence, J. B. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc. Natl Acad. Sci. USA 103, 7688–7693 (2006)
Acknowledgements
We thank V. Boyartchuk, T. Fazzio, E. Heard, P. Kaufman, O. Rando, D. Riethmacher and J. Sharp for advice and/or reagents, D. Kim for help in ES cell analysis, and J. Zhu for statistics. I.B. is a member of the University of Massachusetts DERC (DK32520). This work was supported by National Institutes of Health grants R01CA131158 (National Cancer Institute) and 5 P30 DK32520 (National Institute of Diabetes and Digestive and Kidney Diseases) to I.B., and GM053234 to J.B.L.
Author information
Authors and Affiliations
Contributions
J.S. and I.B. conceived and designed the experiments. M.B., I.H.-B. and I.B. generated the floxed Rnf12 mice. J.S. and Y.C. established and analysed ES cell lines. J.S., M.B., H.M., M.B., N.T.-I., X.Z. and B.J. performed experiments. All authors analysed the data. I.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-14 with legends. (PDF 4592 kb)
Supplementary Movie 1
This movie shows a significant number of cells in the ICM - see Supplementary Information file for full legend. (MOV 2570 kb)
Supplementary Movie 2
This movie shows embryos develops Xist clouds - see Supplementary Information file for full legend. (MOV 6453 kb)
Rights and permissions
About this article
Cite this article
Shin, J., Bossenz, M., Chung, Y. et al. Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice. Nature 467, 977–981 (2010). https://doi.org/10.1038/nature09457
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09457
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.