Abstract
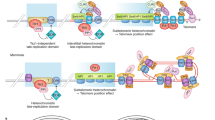
The notion that telomeres are essential for chromosome linearity stems from the existence of two chief dangers: inappropriate DNA damage response (DDR) reactions that mistake natural chromosome ends for double-strand DNA breaks (DSBs), and the progressive loss of DNA from chromosomal termini due to the end replication problem. Telomeres avert the former peril by binding sequence-specific end-protection factors that control the access of DDR activities1,2. The latter threat is tackled by recruiting telomerase, a reverse transcriptase that uses an integral RNA subunit to template the addition of telomere repeats to chromosome ends3. Here we describe an alternative mode of linear chromosome maintenance in which canonical telomeres are superseded by blocks of heterochromatin. We show that in the absence of telomerase, Schizosaccharomyces pombe cells can survive telomere sequence loss by continually amplifying and rearranging heterochromatic sequences. Because the heterochromatin assembly machinery is required for this survival mode, we have termed it ‘HAATI’ (heterochromatin amplification-mediated and telomerase-independent). HAATI uses the canonical end-protection protein Pot1 (ref. 4) and its interacting partner Ccq1 (ref. 5) to preserve chromosome linearity. The data suggest a model in which Ccq1 is recruited by the amplified heterochromatin and provides an anchor for Pot1, which accomplishes its end-protection function in the absence of its cognate DNA-binding sequence. HAATI resembles the chromosome end-maintenance strategy found in Drosophila melanogaster, which lacks specific telomere sequences but nonetheless assembles terminal heterochromatin structures that recruit end-protection factors. These findings reveal a previously unrecognized mode by which cancer cells might escape the requirement for telomerase activation, and offer a tool for studying genomes that sustain unusually high levels of heterochromatinization.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Palm, W. & de Lange, T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 (2008)
Rog, O. & Cooper, J. P. Telomeres in drag: dressing as DNA damage to engage telomerase. Curr. Opin. Genet. Dev. 18, 212–220 (2008)
Blackburn, E. H. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 579, 859–862 (2005)
Baumann, P. & Cech, T. R. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175 (2001)
Miyoshi, T., Kanoh, J., Saito, M. & Ishikawa, F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320, 1341–1344 (2008)
Dunham, M. A., Neumann, A. A., Fasching, C. L. & Reddel, R. R. Telomere maintenance by recombination in human cells. Nature Genet. 26, 447–450 (2000)
Lundblad, V. & Blackburn, E. H. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73, 347–360 (1993)
Teng, S. C. & Zakian, V. A. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae . Mol. Cell. Biol. 19, 8083–8093 (1999)
McEachern, M. J. & Haber, J. E. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75, 111–135 (2006)
Maringele, L. & Lydall, D. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 18, 2663–2675 (2004)
Cooper, J. P., Nimmo, E. R., Allshire, R. C. & Cech, T. R. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385, 744–747 (1997)
Nakamura, T. M., Cooper, J. P. & Cech, T. R. Two modes of survival of fission yeast without telomerase. Science 282, 493–496 (1998)
Wang, X. & Baumann, P. Chromosome fusions following telomere loss are mediated by single-strand annealing. Mol. Cell 31, 463–473 (2008)
Sugawara, N. DNA sequences at the telomeres of the fission yeast S. pombe. PhD thesis, Harvard Univ. (1988)
Toda, T., Nakaseko, Y., Niwa, O. & Yanagida, M. Mapping of rRNA genes by integration of hybrid plasmids in Schizosaccharomyces pombe . Curr. Genet. 8, 93–97 (1984)
Cam, H. P. et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nature Genet. 37, 809–819 (2005)
Kanoh, J., Sadaie, M., Urano, T. & Ishikawa, F. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol. 15, 1808–1819 (2005)
Nakayama, J., Rice, J. C., Strahl, B. D., Allis, C. D. & Grewal, S. I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001)
Ekwall, K. et al. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 109, 2637–2648 (1996)
Nakamura, K. et al. Rad51 suppresses gross chromosomal rearrangement at centromere in Schizosaccharomyces pombe . EMBO J. 27, 3036–3046 (2008)
Sugiyama, T. et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128, 491–504 (2007)
Tomita, K. & Cooper, J. P. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 22, 3461–3474 (2008)
Pitt, C. W. & Cooper, J. P. Pot1 inactivation leads to rampant telomere resection and loss in one cell cycle. Nucleic Acids Res. advance online publication,. 10.1093/nar/gkq580 (3 July 2010)
Ishii, K. et al. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321, 1088–1091 (2008)
Allshire, R. C. & Karpen, G. H. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nature Rev. Genet. 9, 923–937 (2008)
Mason, J. M., Frydrychova, R. C. & Biessmann, H. Drosophila telomeres: an exception providing new insights. Bioessays 30, 25–37 (2008)
Cenci, G., Ciapponi, L. & Gatti, M. The mechanism of telomere protection: a comparison between Drosophila and humans. Chromosoma 114, 135–145 (2005)
Pardue, M. L. & DeBaryshe, P. G. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu. Rev. Genet. 37, 485–511 (2003)
Cenci, G., Siriaco, G., Raffa, G. D., Kellum, R. & Gatti, M. The Drosophila HOAP protein is required for telomere capping. Nature Cell Biol. 5, 82–84 (2003)
Gao, G. et al. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J. 29, 819–829 (2010)
Haering, C. H., Nakamura, T. M., Baumann, P. & Cech, T. R. Analysis of telomerase catalytic subunit mutants in vivo and in vitro in Schizosaccharomyces pombe . Proc. Natl Acad. Sci. USA 97, 6367–6372 (2000)
Moreno, S., Klar, A. & Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe . Methods Enzymol. 194, 795–823 (1991)
Tomita, K. & Cooper, J. P. The telomere bouquet controls the meiotic spindle. Cell 130, 113–126 (2007)
Ferreira, M. G. & Cooper, J. P. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell 7, 55–63 (2001)
Miller, K. M., Rog, O. & Cooper, J. P. Semi-conservative DNA replication through telomeres requires Taz1. Nature 440, 824–828 (2006)
Ferreira, M. G. & Cooper, J. P. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 18, 2249–2254 (2004)
Tomita, K. et al. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol. Cell. Biol. 23, 5186–5197 (2003)
Acknowledgements
We thank T. Cech for discussions and gratefully acknowledge that initial work by T.M.N. on reintroducing Trt1 to circular strains was performed in the Cech laboratory. We thank our current and former laboratory members for discussions and advice. This work was supported by Cancer Research UK.
Author information
Authors and Affiliations
Contributions
D.J. performed the experiments in Figs 3 and 4, Supplementary Figs 6 and 9–17 and Supplementary Tables 1 and 2, and reproduced Figs 1a, b, 2a, d and Supplementary Figs 2 and 8a. A.K.H. first isolated HAATI survivors and performed the experiments in Figs 1 and 2b–d, and Supplementary Figs 2, 4, 5 and 8. T.M.N. performed the experiments in Fig. 2a. K.M.M. first showed that circular strains are hypersensitive to DSB-inducing agents. J.P.C. designed and supervised the study. J.P.C. and D.J. generated the figures and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-17 with legends, Supplementary Tables 1-2 and a Strain Table. (PDF 913 kb)
Rights and permissions
About this article
Cite this article
Jain, D., Hebden, A., Nakamura, T. et al. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature 467, 223–227 (2010). https://doi.org/10.1038/nature09374
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature09374
This article is cited by
-
Telomere and subtelomere high polymorphism might contribute to the specificity of homologous recognition and pairing during meiosis in barley in the context of breeding
BMC Genomics (2023)
-
Heterochromatin replication goes hand in hand with telomere protection
Nature Structural & Molecular Biology (2020)
-
Telomere DNA length-dependent regulation of DNA replication timing at internal late replication origins
Scientific Reports (2019)
-
TASks for subtelomeres: when nucleosome loss and genome instability are favored
Current Genetics (2019)
-
Local enrichment of HP1alpha at telomeres alters their structure and regulation of telomere protection
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.