Abstract
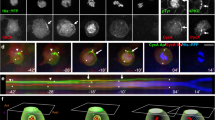
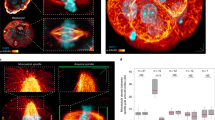
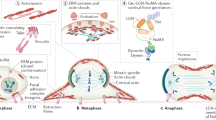
The mitotic spindle determines the cleavage furrow site during metazoan cell division1,2, but whether other mechanisms exist remains unknown. Here we identify a spindle-independent mechanism for cleavage furrow positioning in Drosophila neuroblasts. We show that early and late furrow proteins (Pavarotti, Anillin, and Myosin) are localized to the neuroblast basal cortex at anaphase onset by a Pins cortical polarity pathway, and can induce a basally displaced furrow even in the complete absence of a mitotic spindle. Rotation or displacement of the spindle results in two furrows: an early polarity-induced basal furrow and a later spindle-induced furrow. This spindle-independent cleavage furrow mechanism may be relevant to other highly polarized mitotic cells, such as mammalian neural progenitors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Oliferenko, S., Chew, T. G. & Balasubramanian, M. K. Positioning cytokinesis. Genes Dev. 23, 660–674 (2009)
von Dassow, G. Concurrent cues for cytokinetic furrow induction in animal cells. Trends Cell Biol. 19, 165–173 (2009)
Deng, M., Suraneni, P., Schultz, R. M. & Li, R. The Ran GTPase mediates chromatin signaling to control cortical polarity during polar body extrusion in mouse oocytes. Dev. Cell 12, 301–308 (2007)
Somers, W. G. & Saint, R. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev. Cell 4, 29–39 (2003)
Foe, V. E. & von Dassow, G. Stable and dynamic microtubules coordinately shape the myosin activation zone during cytokinetic furrow formation. J. Cell Biol. 183, 457–470 (2008)
Knoblich, J. A. Mechanisms of asymmetric stem cell division. Cell 132, 583–597 (2008)
Cai, Y. et al. Apical complex genes control mitotic spindle geometry and relative size of daughter cells in Drosophila neuroblast and pI asymmetric divisions. Cell 112, 51–62 (2003)
Fuse, N., Hisata, K., Katzen, A. L. & Matsuzaki, F. Heterotrimeric G proteins regulate daughter cell size asymmetry in Drosophila neuroblast divisions. Curr. Biol. 13, 947–954 (2003)
Albertson, R. & Doe, C. Q. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nature Cell Biol. 5, 166–170 (2003)
Basto, R. et al. Flies without centrioles. Cell 125, 1375–1386 (2006)
Giansanti, M. G., Gatti, M. & Bonaccorsi, S. The role of centrosomes and astral microtubules during asymmetric division of Drosophila neuroblasts. Development 128, 1137–1145 (2001)
Izumi, Y. et al. Differential functions of G protein and Baz-aPKC signaling pathways in Drosophila neuroblast asymmetric division. J. Cell Biol. 164, 729–738 (2004)
Megraw, T. L., Kao, L. R. & Kaufman, T. C. Zygotic development without functional mitotic centrosomes. Curr. Biol. 11, 116–120 (2001)
Field, C. M. & Alberts, B. M. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J. Cell Biol. 131, 165–178 (1995)
Hickson, G. R., Echard, A. & O’Farrell, P. H. Rho-kinase controls cell shape changes during cytokinesis. Curr. Biol. 16, 359–370 (2006)
Minestrini, G., Harley, A. S. & Glover, D. M. Localization of Pavarotti-KLP in living Drosophila embryos suggests roles in reorganizing the cortical cytoskeleton during the mitotic cycle. Mol. Biol. Cell 14, 4028–4038 (2003)
Silverman-Gavrila, R. V., Hales, K. G. & Wilde, A. Anillin-mediated targeting of peanut to pseudocleavage furrows is regulated by the GTPase Ran. Mol. Biol. Cell 19, 3735–3744 (2008)
Royou, A., Sullivan, W. & Karess, R. Cortical recruitment of nonmuscle myosin II in early syncytial Drosophila embryos: its role in nuclear axial expansion and its regulation by Cdc2 activity. J. Cell Biol. 158, 127–137 (2002)
Barros, C. S., Phelps, C. B. & Brand, A. H. Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev. Cell 5, 829–840 (2003)
Giansanti, M. G., Bucciarelli, E., Bonaccorsi, S. & Gatti, M. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 18, 303–309 (2008)
Bonaccorsi, S., Giansanti, M. G. & Gatti, M. Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nature Cell Biol. 2, 54–56 (2000)
Basto, R., Gomes, R. & Karess, R. E. Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila . Nature Cell Biol. 2, 939–943 (2000)
Bowman, S. K. et al. The Drosophila NuMA homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell 10, 731–742 (2006)
Izumi, Y. et al. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nature Cell Biol. 8, 586–593 (2006)
Siller, K. H., Cabernard, C. & Doe, C. Q. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nature Cell Biol. 8, 594–600 (2006)
Conrad, G. W. & Williams, D. C. Polar lobe formation and cytokinesis in fertilized eggs of Ilyanassa obsoleta. I. Ultrastructure and effects of cytochalasin B and colchicine. Dev. Biol. 36, 363–378 (1974)
Kosodo, Y. et al. Cytokinesis of neuroepithelial cells can divide their basal process before anaphase. EMBO J. 27, 3151–3163 (2008)
Siegrist, S. E. & Doe, C. Q. Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development 133, 529–536 (2006)
Cabernard, C. & Doe, C. Q. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila . Dev. Cell 17, 134–141 (2009)
Rolls, M. M. et al. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol. 163, 1089–1098 (2003)
Caussinus, E. & Gonzalez, C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster . Nature Genet. 37, 1125–1129 (2005)
Yu, F. et al. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100, 399–409 (2000)
Woods, D. F. & Bryant, P. J. Molecular cloning of the lethal(1)discs large-1 oncogene of Drosophila . Dev. Biol. 134, 222–235 (1989)
Guan, Z. et al. Mushroom body defect, a gene involved in the control of neuroblast proliferation in Drosophila, encodes a coiled-coil protein. Proc. Natl Acad. Sci. USA 97, 8122–8127 (2000)
Yu, F. et al. Distinct roles of Gαi and Gβ13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J. Cell Biol. 162, 623–633 (2003)
Bonaccorsi, S., Giansanti, M. G. & Gatti, M. Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster . J. Cell Biol. 142, 751–761 (1998)
Buszczak, M. et al. The Carnegie Protein Trap Library: a versatile tool for Drosophila developmental studies. Genetics 175, 1505–1531 (2007)
Martin, A. C., Kaschube, M. & Wieschaus, E. F. Pulsed contractions of an actin-myosin network drive apical constriction. Nature 457, 495–499 (2009)
Dietzl, G. et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila . Nature 448, 151–156 (2007)
Lee, T. & Luo, L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461 (1999)
Siegrist, S. E. & Doe, C. Q. Microtubule-induced Pins/Gαi cortical polarity in Drosophila neuroblasts. Cell 123, 1323–1335 (2005)
Acknowledgements
We thank A. Brand, D. Glover, C.-Y. Lee, M. Peifer, J. Raff, E. Wieschaus, A. Wilde, and the Bloomington and Vienna Drosophila RNAi Center stock centres for fly stocks and/or antibody reagents; R. Andersen, B. Bowerman, M. Goulding and B. Nolan for comments on the manuscript; and T. Gillies and K. Hirono for technical support. This work was supported by the National Institutes of Health (GM068032; to K.P.), the American Heart Association (to C.C. and K.P.), the Swiss National Science Foundation (to C.C.) and HHMI (to C.Q.D.).
Author information
Authors and Affiliations
Contributions
C.C., K.E.P. and C.Q.D. conceived and designed the project. C.C. performed all the experiments. C.C. and C.Q.D. wrote the manuscript with input from K.E.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-4 with legends and legends for Supplementary Movies 1-11. (PDF 310 kb)
Supplementary Movies 1-3
This movie shows that Pavarotti, Anillin and Myosin is localized to the larval neuroblast basal cortex in early anaphase C (see Supplementary Information file for full legend). (MOV 3243 kb)
Supplementary Movie 4
This movie shows that asymmetric Myosin localization does not require the mitotic spindle (see Supplementary Information file for full legend). (MOV 959 kb)
Supplementary Movie 5
This movie shows that an apically displaced spindle does not alter the asymmetric localization of Myosin (see Supplementary Information file for full legend). (MOV 1384 kb)
Supplementary Movie 6
This movie shows that the altered spindle orientation does not change Pavarotti basal localization (see Supplementary Information file for full legend). (MOV 932 kb)
Supplementary Movie 7
This movie shows that the altered spindle orientation does not change Myosin localization to the basal cortex (see Supplementary Information file for full legend). (MOV 610 kb)
Supplementary Movie 8
This movie shows that the altered spindle orientation results in the formation of basal polar lobes (see Supplementary Information file for full legend). (MOV 2661 kb)
Supplementary Movie 9
This movie shows that polarity-induced furrow initiation precedes spindle-induced furrow initiation (see Supplementary Information file for full legend). (MOV 375 kb)
Supplementary Movie 10
This movie shows that pins are required for polarity-induced furrow formation during symmetric cell divisions (see Supplementary Information file for full legend). (MOV 419 kb)
Supplementary Movie 11
This move shows that Basal Myosin localization is independent of spindle symmetry (see Supplementary Information file for full legend). (MOV 1497 kb)
Rights and permissions
About this article
Cite this article
Cabernard, C., Prehoda, K. & Doe, C. A spindle-independent cleavage furrow positioning pathway. Nature 467, 91–94 (2010). https://doi.org/10.1038/nature09334
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature09334
This article is cited by
-
Two RhoGEF isoforms with distinct localisation control furrow position during asymmetric cell division
Nature Communications (2023)
-
Polarized branched Actin modulates cortical mechanics to produce unequal-size daughters during asymmetric division
Nature Cell Biology (2023)
-
Asymmetric chromatin retention and nuclear envelopes separate chromosomes in fused cells in vivo
Communications Biology (2022)
-
Wnt signaling polarizes cortical actin polymerization to increase daughter cell asymmetry
Cell Discovery (2022)
-
Diffusive Interface Model for Actomyosin Driven Cell Oscillations
Bulletin of Mathematical Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.